INTRODUCTION
How cell fate specification is achieved during animal embryogenesis is a fundamental issue in developmental biology. In ascidian embryos, fibroblast growth factor (FGF) 9/16/20 signaling induces specification of notochord, mesenchyme, and brain cells at the early embryonic stages. Notochord and mesenchyme are induced in the marginal zone of the vegetal hemisphere by a FGF9/16/20 signal emanating from endodermal cells, as in vertebrate embryos (Nakatani et al., 1996; Kim et al., 2000; Imai et al., 2002; Kumano et al., 2006). When FGF signaling was inhibited using inhibitors of FGF/MEK/Ras/Erk (extracellular signal-regulated kinases; MAP kinases) signaling, the expression of differentiation markers was not detected in mesenchyme and notochord precursor cells. The FGF9/16/20 signal also induces the a-line neural cells and suppresses epidermal fate in ascidians (Inazawa et al., 1998; Bertrand et al., 2003). Fgf9/16/20 morpholino oligo suppresses expression of the Otx brain marker. Inhibition of Ras/MEK/Erk/Ets signaling leads to loss of the expression of Otx and HrETR1 neural markers in a-line cells (Kim & Nishida, 2001; Miya & Nishida, 2003). Ets (E-twenty six; E26 transformation-specific) and GATA transcription factors are activated by FGF9/16/20 signal in a-line cells. Ets and GATA in response to the FGF signal, activates the expression of Otx only in the animal hemisphere, which leads to brain formation (Bertrand et al., 2003). Ets is also activated in cells of the vegetal hemisphere and is required for notochord and mesenchyme specification (Miya & Nishida, 2003).
The Ets family members are target transcription factors of the Ras/MEK/Erk signaling and involved in a wide range of processes, tumorigenesis, proliferation, lymphoid differentiation, and developmental regulation (Bartel et al., 2000; Yordy & Muise-Helmericks, 2000; Tootle & Rebay, 2005). They have the common winged-helix DNA binding domain. Most of the known Ets family proteins have been shown to activate transcription. However, several Ets proteins like Tel, Net and Erf subfamilies can act as transcriptional repressors. It was reported that Erf (Ets2 repressor factor) is also regulated by the Ras/Erk signaling pathway (Le Gallic et al., 1999; Twigg et al., 2013). In mammal, extraembryonic ectoderm differentiation requires the nuclear localization and function of Erf proteins with attenuation of FGF/Erk signaling (Papadaki et al., 2007). Erf contributes to the proper trophoblast differentiation, as an effector in the FGF signaling. However, it has not been known whether Erf is involved in FGF/Ras/Erk/Ets signaling pathway of ascidian embryos.
In this study, in order to determine Erf function in ascidian embryos, we isolated and characterized the ascidian Erf gene. Its expression is observed in mesenchyme and a-line neural precursor cells at the gastrula stage. The MEK/Erk signaling-inhibited embryos showed loss of Erf expression in mesenchyme cells, and excess expression of Erf in a-line neural cells.
MATERIALS AND METHODS
Adults of the ascidian Halocynthia roretzi were collected near the Marine Biology Center for Research and Education, Gangneung, Korea. Naturally spawned eggs were fertilized with a suspension of sperm from another individual and cultured in filtered seawater containing 50 μg/ml streptomycin sulfate and 50 μg/ml kanamycin sulfate at 9–13°C. Embryos were collected at appropriate stages and fixed for whole-mount in situ hybridization. Tissues dissected from adults immediately frozen in liquid nitrogen and stored at –80°C until RNA extraction.
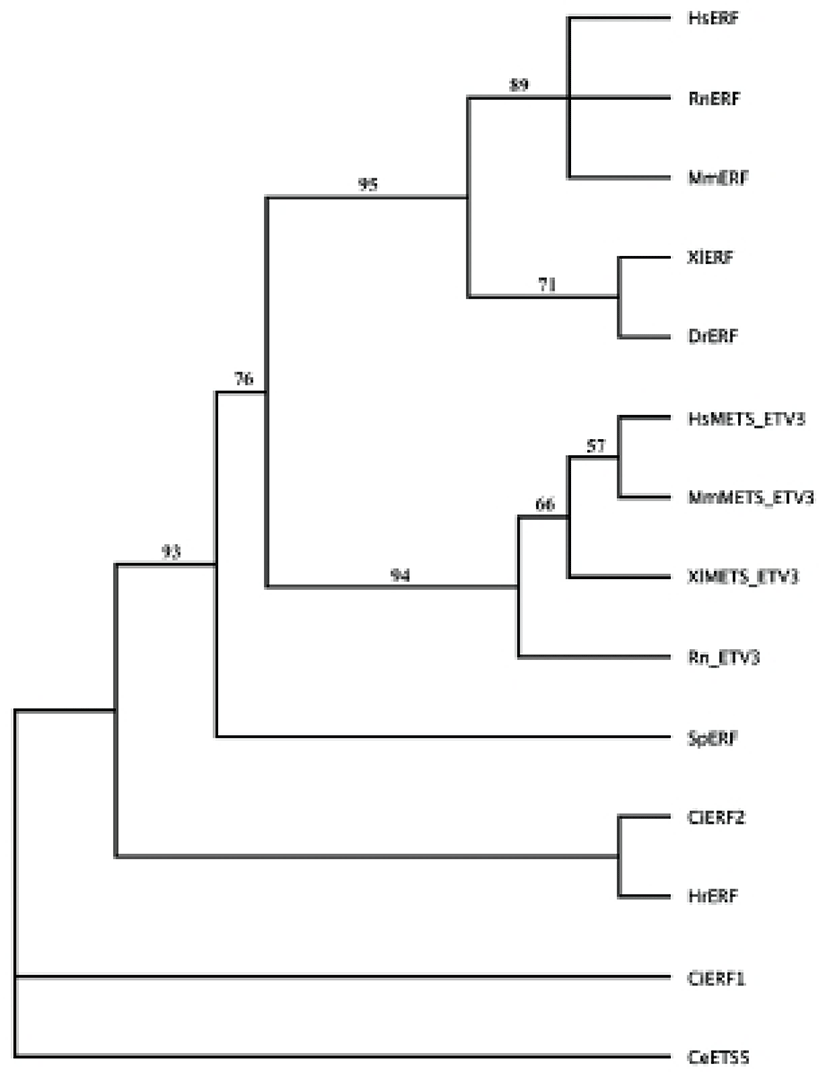
A cDNA fragment for Erf homolog was found in clones of the database of maternal mRNA of H. roretzi eggs, MAGEST (Kawashima et al., 2000; Lee et al., 2011). In order to obtain a full-length H. roretzi Erf cDNA, the first strand cDNA was synthesized from extracted mRNA using SMART RACE cDNA synthesis kit (Clontech). The Erf homologue was named Hr-Erf (H. roretzi Erf). The amino acid sequences of Erf gene products were aligned on the basis of maximum similarity using Clustal W program. Molecular phylogenetic relationships among the Erf and Mets/Etv gene products were estimated using the neighbor-joining method (Saitou & Nei, 1987). Sequence data used in this study were taken from GenBank/EMBL/NCBI databases, with following accession numbers: CiERF1, Ciona intestinalis ERF1 (XP002124625); CiERF2, Ciona intestinalis ERF2 (NP001071696); DrERF, Danio rerio ERF (NP001038392); HsERF, Homo sapiens ERF/PE2 (NP006485); MmERF, Mus musculus ERF (NP034285); RnERF, Rattus norvegicus ERF (NP001163806); XlERF, Xenopus laevis ERF (NP001089722); SpERF, Strongylocentrotus purpuratus ERF (XP800545); HsMETS, Homo sapiens METS/ETV3/PE1 (NP001138784); MmMETS, Mus musculus METS/Etv3/PE1 (NP001076787); RnETV, Rattus norvegicus ETV3 (NP001099920); XlMETS, Xenopus laevis METS/ETV3 (NP001088435); CeETS5, Caenorhabditis elegans ETS5 (NP508865).
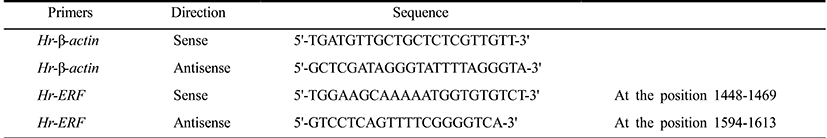
Total RNAs were extracted from fertilized eggs and embryos at various developmental stages using the RNeasy kit (Qiagen) and each RNA (0.5 μg) was reverse-transcribed to first strand cDNA using M-MLV reverse transcriptase (Bioneer). QPCR was performed in a volume of 20 μl containing the synthesized cDNA (0.3 μg each), the 2× SYBR Premix Ex Taq (TaKaRa), the 50× ROX Reference Dye II (TaKaRa), and 10 μM primer set (Table 1) designed by Primer Express v3.0 software (Applied Biosystems) on a QPCR System (Applied Biosystems 7500). Thermal cycling was performed with a two-step PCR protocol: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 5 sec and 60°C for 34 sec. Data were represented by the mean ± S.E.M. of three independent samples. Relative quantitation values were expressed using the 2–ΔΔCt method as fold changes in the Erf mRNA normalized to β-actin mRNA of H. roretzi.
To inhibit the MEK/Erk signaling, embryos were treated with 2 μM U0126 (Promega) at the early 32-cell stage for 20 min (Kim & Nishida, 2001; Lee et al., 2011). U0126 is an MEK inhibitor that inhibits both the activation of MEK and Erk (MAPK). U0126 was dissolved in dimethylsulfoxide at a concentration of 10 mM and diluted with seawater to the final concentration just before use.
To examine the expression patterns of Hr-Erf, whole-mount in situ hybridization was performed according to the method of Miya et al. (1997) with minor modification. Hr-Erf antisense probes for in situ hybridization were prepared with a digoxigenin RNA labeling kit (Roche).
RESULTS AND DISCUSSION
To elucidate interactions of transcription factors involved in the FGF/Ras/Erk/Ets signaling pathway, we isolated Hr-Erf, an ascidian H. roretzi orthologue of vertebrate Erf. The Hr-Erf cDNA encompassed 3110 nucleotides including a potential signal sequence for polyadenylation at the 3' end and encoded a predicted polypeptide of 760 amino acids (Fig. 1). In the nucleotide, the ATG at the position 216-218 represents the putative start codon and the asterisk at the position 2496-2498 indicates the termination codon. The deduced polypeptide had the Ets DNA-binding domain in its N-terminal region (Fig. 1). The overall degree of amino acid identity between the Hr-Erf and vertebrate Erf genes was approximately 30%, however, the Ets domain was highly conserved (~80%; data not shown). A characteristic feature of Ets family is that they shear an evolutionarily-conserved Ets domain of about 90 amino acids which binds purine-rich DNA sequences centered over a GGAA/T core motif (Sgouras et al., 1995; Oikawa & Yamada, 2003). Most of Ets family proteins have the Ets domains in their C-terminal regions, whereas several Ets subfamilies including Erf and TCF (ternary complex factor) possess the Ets domains in their N-terminal regions. We could not decide position of the repression domain in the C-terminal region of Hr-Erf. It was reported that the repression domain scarcely has similarities between the vertebrate Erf proteins (Mavrothalassitis & Ghysdael, 2000). To examine the relationship of Hr-Erf with various Erf/METS/ETV3 proteins, we assembled a molecular phylogenetic tree by use of the Ets domains (Fig. 2). The phylogenetic tree clearly showed that the Hr-Erf is the Halocynthia orthologue of vertebrate Erf genes. METS/ETV3 (PE1) is a related member of the Erf family of transcriptional repressors (Oikawa & Yamada, 2003).

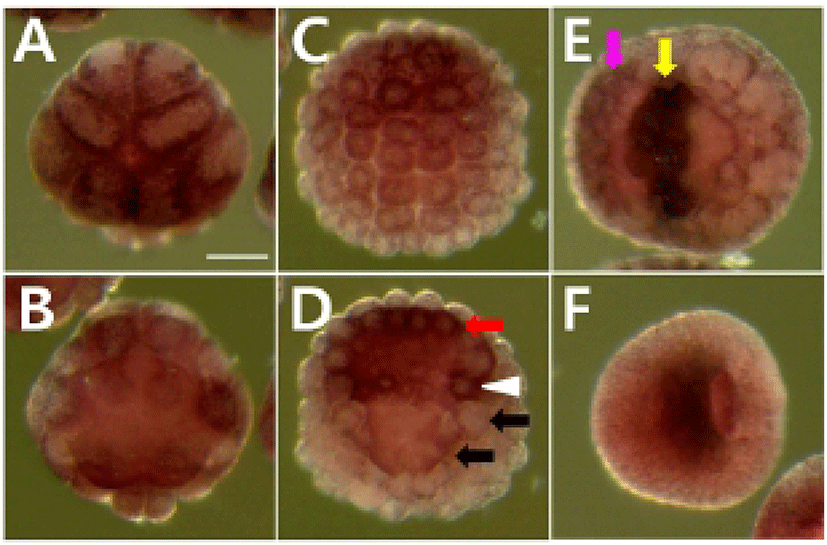
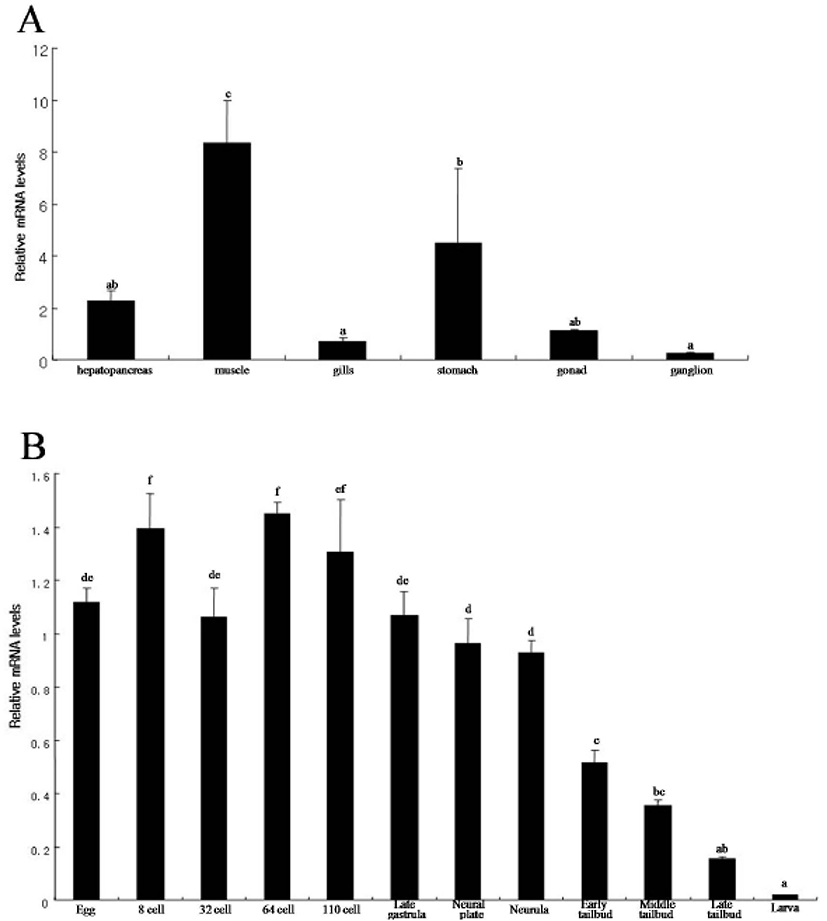
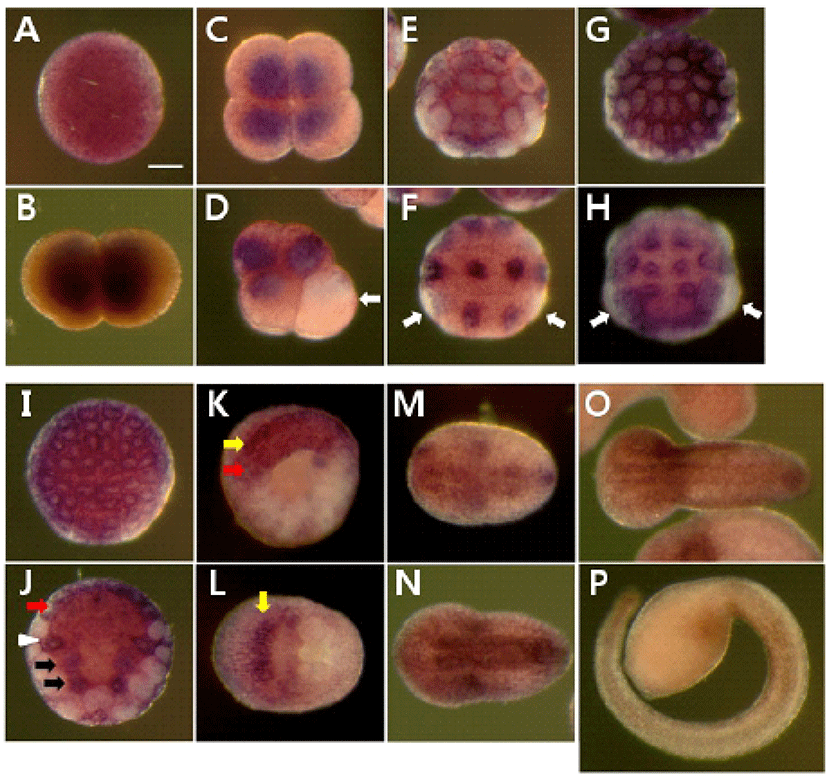
We determined the expression pattern of Hr-Erf mRNA in various adult tissues and embryos using quantitative real-time PCR (QPCR). In adult animals, Hr-Erf mRNA was predominantly detected in the muscle, and at lower levels in ganglion, gills, gonad, hepatopancreas, and stomach (Fig. 3A). During embryogenesis, Hr-Erf mRNA was detected from eggs to early developmental stage embryos, whereas the transcript levels were decreased from the early tailbud stage (Fig. 3B). Increasing mRNA level at the 8-cell stage indicates zygotic expression of Hr-Erf. To further examine the spatial expression of Hr-Erf mRNA, we carried out whole-mount in situ hybridization at various stages. Similar to the QPCR results, a significant amount of maternal transcripts of Hr-Erf was evenly distributed in the cytoplasm of fertilized eggs (Fig. 4A). The maternal transcripts were restricted to the animal pole cells from the 8-cell to gastrula stages (Fig. 4D-I). Maternal transcripts of Hr-Erf were gradually lost from the late gastrula stage (Figs. 4I, K; 3B). Zygotic expression of Hr-Erf started in most blastomeres at the 8-cell stage, when its signal was evident in their nuclei, except for the B4.1 blastomeres (Figs. 4D; 3B). At gastrula stage, Hr-Erf was expressed in the precursor cells of mesenchyme (Fig. 4J, black arrows), nerve cord (Fig. 4J, red arrow) and brain (Fig. 4K, yellow arrow) as well as in trunk lateral cells (Fig. 4J, white arrowhead). The expression was detected exclusively in brain precursor cells at the neural plate stage (Fig. 4L, yellow arrow). These results suggest that zygotic Hr-Erf products are involved in specification of brain and mesenchyme cells. There is a conspicuous feature that the Hr-Erf expression was suppressed in the B4.1 cells (Fig. 4D, white arrow) and in their muscleline descendants (Fig. 4F, H, white arrows). The B4.1 cells give rise to mesenchyme and endoderm as well as muscle (Kim et al., 2000). Except in muscle precursors, expression of Hr-Erf was started newly in other descendants of the B4.1 cells at the next stages (Fig. 4F, H, J). This finding suggests that suppression of Hr-Erf expression is required for muscle cell formation. Erf is ubiquitously expressed throughout mouse embryonic development and adulthood (Papadaki et al., 2007). In the developing placenta, Erf expression is restricted to the extraembryonic ectoderm after 7.5 days postcoitum. Its expression is restricted in a subpopulation of labyrinth cells after 9.5 days postcoitum. Sp-Erf, a sea urchin Strongylocentrotus purpuratus orthologue of vertebrate Erf, is ubiquitously expressed during cleavage stages, but slowly decreases until early blastula (Rizzo et al., 2006). Sp-Erf expression level increases insignificantly again during gastrulation. Its expression is detected dominantly in endoderm and oral ectoderm in gastrula. In these animal embryos, Erf expression is presented from fertilized eggs to gastrulae like Hr-Erf.
To analyze whether the MEK/Erk signaling is involved in regulation of Hr-Erf expression, we examined the expression of Hr-Erf in embryos treated with MEK inhibitor, U0126. U0126 inhibits both the activation of MEK and Erk in ascidian embryos (Kim & Nishida, 2001). Maternal expression of Hr-Erf was normally detected in animal cells in the MEK signaling-blocked embryos (Fig. 5A). MEK inhibitor did not affect the Hr-Erf expression in trunk lateral cells and nerve cord precursors at the gastrula stage (Fig. 5D, white arrowhead and red arrow, respectively). The MEK inhibitor-treated embryos showed downregulation of Hr-Erf in the mesenchyme precursors (Fig. 5D, black arrows), whereas they displayed increase of Hr-Erf expression in the a-line neural precursor cells (Fig. 5E, yellow arrow). Interestingly, the MEK signaling-inhibited embryos had ectopic expression of Hr-Erf in the a-line peripheral neural precursors at the neural plate stage (Fig. 5E, pink arrow). These results suggest that MEK/Erk signaling is required for the expression of Hr-Erf in the mesenchyme and a-line neural cells. Erf is an transcriptional repressor that is regulated by Erk signal via subcellular localization (Sgouras et al., 1995). It was also reported that mammalian Erf locates nuclear and can suppress cell division cycle in the absence of Erk activity (Le Gallic et al., 1999; Papadaki et al., 2007). It is unclear how Hr-Erf function is regulated in ascidian embryonic development. This mechanism should be focused on in future studies.