INTRODUCTION
Myogenesis (or muscle development) is composed of the specification processes of the individual myogenic progenitor cells according to myogenic lineage, proliferation and migration, differentiation, and fusion. The various myogenic regulatory factors (MRFs) are involved in the commitment of mesodermal cells to a muscle lineage (i.e., MyoD, Myf5) and in the initiation and maintenance of the terminal differentiation program (i.e., Myogenin, Mrf4) (Arnold & Braun, 1996). MRFs are highly conserved in teleost and mammals and whose mutation will result in muscular defects (Hasty et al., 1993; Megeney et al., 1996; Rescan, 2001; Kassar-Duchossoy et al., 2004). Although there are several common features in muscle development between teleost and mammals, myogenesis in teleost has some unique features compared to mammals, which include the early stage of muscle commitment, presence of adaxial cells, different proportions of slow and fast fibers, and muscle growth throughout much of ontogeny (Rossi & Messina, 2014). Whereas muscle development in mammals is conventionally divided into pre-natal and post-natal, the main phases of myogenesis in teleost consist of the embryonic-larval-juvenile and adult stages (Johnston et al., 1998; Rossi & Messina, 2014).
The LUC7-like gene (Luc7l) encodes a putative RNA-binding protein similar to the yeast Luc7p subunit of the U1 snRNP splicing complex, which has a role in 5' splice site recognition (Fortes et al., 1999). The LUC7L is serinearginine- rich protein (SR protein) that localizes in the nucleus through its arginine-serine-rich domain (RS domain) at the C-terminus (Kimura et al., 2004). LUC7L homologs were identified in other species including human Homo sapiens (GenBank accession no. NP_958815.1), red junglefowl Gallus gallus (XP_414651.3), painted turtle Chrysemys picta bellii (XP_005294482) and zebrafish Danio rerio (XP_005162951). Human LUC7L, also called SR+89 or putative SR protein LUC7B1, is closely related to cisplatin resistance-associated over-expressed protein (CROP), which makes anticancer therapy failed (Nishii et al., 2000). Mouse LUC7L plays an important role in the regulation of muscle differentiation (Kimura et al., 2004). LUC7L expression is negatively regulated during the course of development of limb skeletal muscle and during in vitro differentiation of the mouse myoblast cell lines (Kimura et al., 2004). However, the identification and characterization of LUC7L remains to be elucidated in many species.
Korean rose bitterling (Rhodeus uyekii) belongs to the Acheilognathinae subfamily of the Cyprinidae family. It is a common freshwater fish endemic to Korea found in rivers that empty into the Western and Southern Sea of Korea (Kong et al., 2012). This species has been proposed as a candidate for developing ornamental fish because of its small size and beautiful body color (Kang et al., 2005). Genetic studies on the Korean rose bitterling reported the complete mitochondrial genome sequence of R. uyekii (Kim et al., 2006) and development of microsatellite makers for evaluation of population genetic diversity (Kim et al., 2014). The R. uyekii β-actin gene has been suggested as a promoter capable of driving constitutive transgene expression (Kong et al., 2014). In this study, we report the identification and molecular characterization of the Luc7l cDNA of Korean rose bitterling (RuLuc7l). We analyzed multiple alignments of the deduced RuLUC7L polypeptide sequence and other LUC7L homologs. We investigated the expression of RuLuc7l transcript during early development of Korean rose bitterling and in several tissues of Korean rose bitterling. This study is the first report of molecular and functional analyses of the Korean rose bitterling Luc7l gene.
MATERIALS AND METHODS
The RuLuc7l cDNA sequence was isolated from the EST analysis of the Korean rose bittering R. uyekii cDNA library (data not shown). EST clones were isolated from the R. uyekii cDNA library using a Plasmid Miniprep Kit (Qiagen), and sequenced using T3 reverse primers (Promega) and an ABI3730xl automatic sequencer (Applied Biosystems). Based on partial sequence sequenced, EST clones were sequenced using designated internal primers (RuLuc7lseq 1, 5'-CCT ACT TGG GCC TCC ATG ATA-3'; RuLuc7l-seq 2, 5'-ACA GAG AGG CGG GAG AGA TC- 3'). The nucleotide sequence was analyzed and compared using the BLASTX search program (http://www.ncbilnlm.nih.gov/BLAST/) .
The relevant sequences were compared using the BLASTX search program (http://www.ncbi.nlm.nih.gov/BLAST/) and retrieved from GenBank for multiple sequence alignments using CLUSTALW (http://www.genome.jp/tools-bin/clustalw). MEGA (ver. 4) was used to assess similarities among the aligned sequences. A phylogenetic tree based on the deduced amino acid sequences was constructed using a neighbor-joining algorithm, and the reliability of the branching was tested using bootstrap resampling with 1,000 pseudo-replicates.
Total RNA was prepared from tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions, treated with DNase I (New England BioLabs, Beverly, MA, USA) and quantitatively determined; 500 ng samples were used for reverse transctiption (RT). First-strand cDNA was synthesized using Transcriptor First Strand cDNA Synthesis Kit (Roche). Quantitative real-time PCR was performed using Fast SYBR Green Master Mix (Applied Biosystems, Inc.) and the following forward and reverse primers : RuLuc7l, RuLuc7l-RT-F (5'-TGG GCC TCC ATG ATA ACG A-3') and RuLuc7l-RT-R (5'-GAA GCC CAA GTG CAG TTT GC-3'); and Ruβ-actin (GenBank accession no. JQ279058), RubAct-RT-F (5'-GAT TCG CTG GAG ATG ATG CT-3') and RubAct-RT-R (5'-ATA CCG TGC TCA ATG GGG TA-3'). Following an initial 10-min Taq activation step at 95°C, real-time PCR was performed using the following cycling conditions: 40 cycles of 95°C for 10 s, 60°C for 15 s, and fluorescence reading in an SDS 7500 system (Applied Biosystems, Inc.). Transcript levels were quantified as expression relative to the β-actin transcript level.
R. uyekii were collected from the Yangchun River, Uiryung-gun, Gyungnam, Republic of Korea. The fish were maintained at the National Fisheries Research and Development Institute (NFRDI) in Busan, Republic of Korea. The adults were maintained in 40 L glass aquaria at a density of approximately 20 fish per aquarium. The water was renewed weekly and the temperature in the rearing tanks was maintained at 20 ± 1°C. The room was maintained on a 12:12-h light:dark cycle. Adults were fed TetraBits (Tetra) and frozen bloodworms (Advanced Hatchery Technology) twice a day. For RNA extraction, tissues were removed from three R. uyekii (mean body weight: 0.75 ± 0.29 g; mean total length: 4.0 ± 0.23 cm), immediately frozen in liquid nitrogen, and kept separately and stored at –80°C before use.
RESULTS
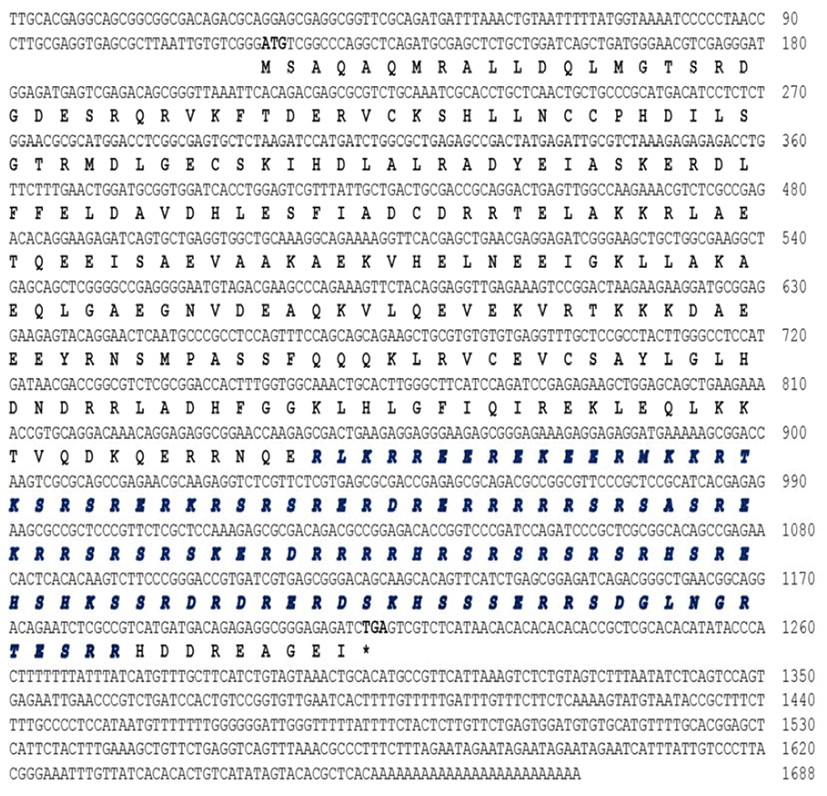
The RuLuc7l cDNA sequence was identified from the expressed sequence tag (EST) analysis of the Korean rose bittering R. uyekii cDNA library. The EST clone, RU-2- 4a_D22, of 1688 bp contains 1095-nt ORF encoding a 364-aa protein, preceded by a 120-nt 5' UTR, and followed by a 473-nt 3' UTR with a poly(A) tail (Fig. 1). A search using the BLASTP program (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) revealed that the deduced amino acids of RuLuc7l have a serine/arginine-rich region (amino acids R243-R355), which is found commonly in other Luc7l proteins.
The deduced amino acids of RuLuc7l were aligned with LUC7L proteins from other species including human using GENETYX software. Pairwise alignment revealed RuLUC7L showed high amino acid identities of 71% - 97% with those of other species, including fish, birds, mammals, amphibians and reptiles. Multiple alignments revealed that RuLUC7L shared the high homology with other LUC7L proteins in the N-terminal region, but relatively low in the C-terminal region despite of the presence of serine/argininerich region (Fig. 2).

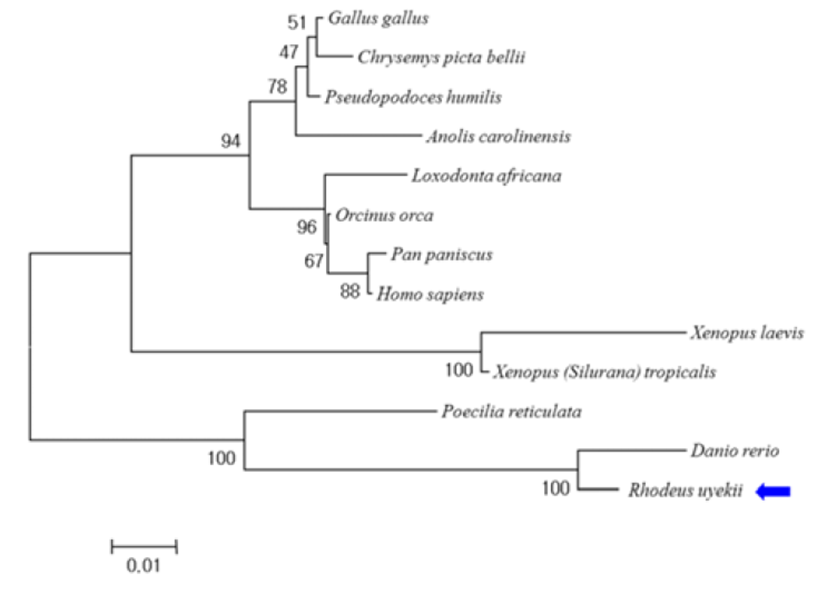
A phylogenetic analysis was performed based on the deduced amino acid sequence of RuLuc7l and related sequences. The tree indicated clear clustering of LUC7L sequences into two groups: those from Korean rose bitterling, zebrafish, guppy, african bush elephant, green anole, bonobo, killer whale, ground tit, red jungle fowl, painted turtle, western clawed frog, african clawed frog and human. RuLUC7L formed a cluster with LUC7Ls from zebrafish and guppy, and was phylogenetically separate from other species (Fig. 3).
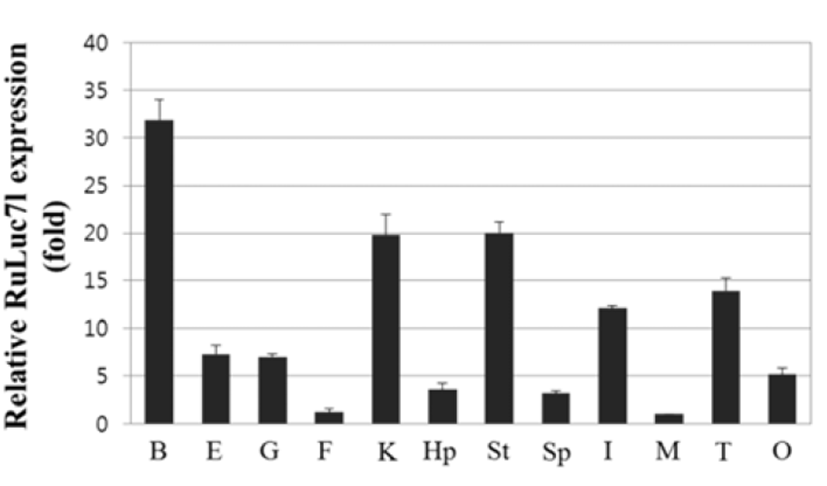
Quantitiative real-time PCR was performed to examine the tissue distribution of RuLuc7l mRNA. The expression levels of RuLuc7l mRNA were quantified after normalization to β-actin as an internal reference gene. Expression of RuLuc7l mRNA was detected in all tissue examined; highly in the brain, stomach and kidney and moderately in the intestine and testis. Levels of the RuLuc7l mRNA in the brain, stomach and kidney were 31.8, 20.0 and 19.8 folds that in muscle where expression was the lowest, respectively (Fig. 4).
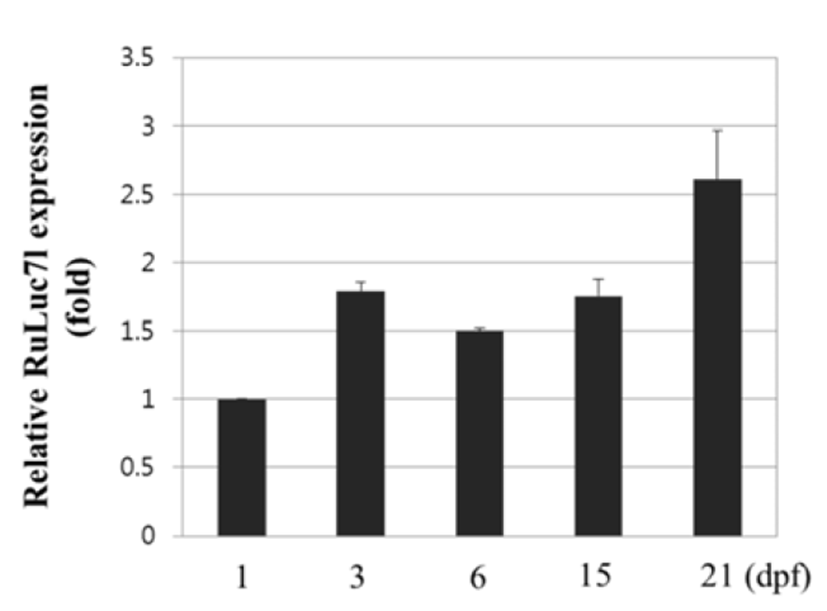
The expression of RuLuc7l mRNA during early development of Korean rose bitterling was determined by quantitative real-time PCR at 1, 3, 6, 15 and 21 days postfertilization (dpf). The expression of RuLuc7l mRNA was detected from 1 dpf and moderately increased until 21 dpf during the early development. Levels of the RuLuc7l mRNA at 21 dpf was 2.61 folds that at 1 dpf (Fig. 5).
DISCUSSION
We previously constructed a cDNA library of the Korean rose bitterling R. uyekii and carried out EST analysis of approximately 1,000 clones. One of the EST clones contained an open reading frame encoding 364 amino acids which is showing 71-97% sequence homology with Luc7ls of other species; zebrafish, guppy, african bush elephant, green anole, bonobo, killer whale, ground tit, red jungle fowl, painted turtle, western clawed frog, african clawed frog and human. The deduced amino acids of RuLuc7l had the serine/arginine-rich region conserved with those of other species. LUC7L belongs to serine/arginine (SR) proteins, characterized by a C-terminal RS domain (Kimura et al., 2004). SR proteins are a conserved protein families involved in RNA splicing, which are commonly found in the nucleus (Zahler et al., 1992). A gene trap mutant protein partially missing its RS domain, Luc7lGT, lost speckled distribution in the nucleus, suggesting the RS domain is also important for the intranuclear distribution of LUC7L (Kimura et al., 2004). The presence of RS domain in RuLUC7L suggests that RuLUC7L possibly localizes in the nucleus and might play a role as a regulatory factor in RNA splicing in Korean rose bitterling.
Transcript of RuLuc7l was ubiquitously expressed in all tissue examined and highly expressed in the brain, stomach and kidney. During the early development of Korean rose bitterling, the expression of RuLuc7l was detected after fertilization and increased until 21 dpf. This expression pattern is consistent with previous results. Mouse Luc7l was detected in the brain, liver, testis, spleen, and even in the embryo, but not in the skeletal muscle (Kimura et al., 2004). Because LUC7L as an RNA splicing factor is involved in gene expression and regulation (O’Reilly et al., 2013), RuLuc7l might be suggested to be increased during early development. Recent study demonstrated that forced expression of LUC7L protein regulated myogenesis in vitro (Kimura et al., 2004). The level of RuLuc7l mRNA in the muscle was low compared to those of other tissues. These results suggest that RuLUC7L may play a role in the development in a variety of tissue like myogenic regulation.
Cisplatin resistance-associated overexpressed protein (CROP) is the human homologue of yeast LUC7P (Umehara et al., 2003). CROP is a novel putative SR protein sharing the common function with LUC7L. CROP/hLuc7A has been studied as a key molecule of cisplatin resistance of anticancer therapy targeting a variety of malignant tumors because it is overexpressed in cisplatin-resistant cell lines (Umehara et al., 2003). In this context, LUC7L is considered as a possible target for drug resistance in tumor in a manner similar to its mammalian counterparts.
In the present study, we have done the molecular characterization and functional analysis of Luc7l of Korean rose bitterling R. uyekii. Sequence and homology analyses showed that the deduced amino acid sequence of RuLuc7l has been evolutionally conserved. Also, its tissue distribution and expression level during early development provide clues for the function involved in gene regulation and development. For understanding biological activity of LUC7L, further studies are required to elucidate the developmental functions of RuLUC7L in myogenesis in R. uyekii.

