ARTICLE
Differences and Variations among Anguilla japonica, Muraenesox cinereus and Conger myriaster from the Yellow Sea
Jong-Man Yoon†
Author Information & Copyright ▼
Dept. of Aquatic Life Medicine, College of Ocean Science and Technology, Kunsan National University, Gunsan 573-701, Korea
†Corresponding author : Jong-Man Yoon, Dept. of Aquatic Life Medicine, College of Ocean Science and Technology, Kunsan National University, Gunsan 573-701, Korea. Tel. : +82-63-469-1887,
jmyoon@kunsan.ac.kr
Copyright © 2015 © The Korean Society of Developmental Biology, All rights reserved. This is an Open-Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received: Jul 6, 2015 ; Revised: Jul 16, 2015 ; Accepted: Aug 3, 2015
ABSTRACT
Genomic DNAs were extracted from the muscle of twenty-one specimens of three eel species collected in Anguilla japonica (AJ), Muraenesox cinereus (MC) and Conger myriaster (CM) from the Yellow Sea, respectively. In the present study, 7 oligonucleotides primers generated 191 specific loci in the AJ species, 226 in the (MC) species and 181 in the CM species, respectively. The primer BION-02 generated the most loci (a total of 83), with an average of 11.86 in the AJ species. The specific loci generated by oligonucleotides primers exhibited inter-individual-specific characteristics, thus revealing DNA polymorphisms. With regard to average bandsharing value (BS) results, individuals from Conger myriaster species (0.808) exhibited higher bandsharing values than did individuals from Muraenesox cinereus species (0.729) (p<0.05). The longest genetic distance (0.430) displaying significant molecular difference was also between individual no. 01 within Anguilla japonica eel species and individual no. 04 within Anguilla japonica species. In this study, the dendrogram resulted from reliable seven oligonucleotides primers, indicating three genetic clusters composed of group I (ANGUILLA 01~ANGUILLA 07), group II (MURAENESOX 08~MURAENESOX 14) and group III (CONGER 15~CONGER 21). The existence of species differentiation and DNA polymorphisms among three eel species were detected by PCR analysis. As mentioned above, a dendrogram revealed close relationships between individual identities within three eel species. High levels of a significant genetic distance among three eel species showed this PCR approach is one of the most suitable tools for individuals and/or species biological DNA studies.
Keywords: Anguilla japonica; Conger myriaster; Eel species; Genetic distance; Muraenesox cinereus
INTRODUCTION
Three eel species such as Anguilla japonica (AJ), Muraenesox cinereus (MC) and Conger myriaster (CM), belonging to the order Anguilliformes, are the most popular marine products in Korea because of their taste and nutritional value, and Koreans consume them in large quantities. Eel, ecologically important warm water fish species widely distributed on the coast of the Yellow Sea, southern sea and the several sea areas under the natural ecosystem. As the eel culture industry grows, so doe’s interest into the genetics of this finfish species. However, in spite of their economic and scientific consequences, a little information currently exists regarding the genetic levels only of eel species in Korea and/or the other countries (Min & Yang, 1993; Jing & Li, 1999; Kimura et al., 2004; Oh, 2011). In this study, to explicate the genetic distances and differences among geographical eel species, the author accomplished a clustering analysis of three eel species collected from the Yellow Sea.
MATERIALS AND METHODS
PCR analysis was performed on DNA samples extracted from a total of 21 individuals using seven oligonucleotides primers. Muscle tissues were obtained separately from individuals from Anguilla japonica, Muraenesox cinereus and Conger myriaster, respectively. Eel muscle was collected in sterile tubes, instantaneously placed in liquid nitrogen, and stored at –40°C until the genomic DNA extraction. Genomic DNA was extracted and purified under the conditions described previously (Kim et al., 2006; Yoon, 2008). After several washings, lysis buffer I (155 mM NH4Cl; 10 mM KHCO3; 1 mM EDTA) was added to the samples, and the mixture tubes were gently inverted. The concentration of the extracted genomic DNA was measured by optical density at 260 nm by a spectrophotometer (Beckman Coulter, Buckinghamshire, UK). PCR was performed using two Programmable DNA Thermal Cyclers (MJ Research Inc., Waltham, MA, USA). Seven oligonucleotides primers such as BION-02 (5’-CAATCGCCGT- 3’), BION-14 (5’-TGGATTGGTC-3’), BION-22 (5’-GGT ACTCCAC-3’), BION-30 (5’-GCCACCTCCT-3’), BION- 34 (5’-GCCGCTACTA-3’), BION-35 (5’-AGCGGCTAGG- 3’) and BION-40 (5’-GTTGCGATCC-3’) were displayed to generate the shared loci, specific loci, unique shared loci to each species and shared loci by the three species which could be obviously scored. The 100-bp DNA ladder (Bioneer Corp., Daejeon, Korea) was used as the DNA molecular weight marker. The electrophoresed agarose gels were illuminated by ultraviolet rays, and photographed using a photoman direct copy system. Euclidean genetic distances within- and between-species were also calculated using the hierarchical dendrogram program Systat ver.10 (SPSS Inc., Chicago, IL, USA).
RESULTS AND DISCUSSION
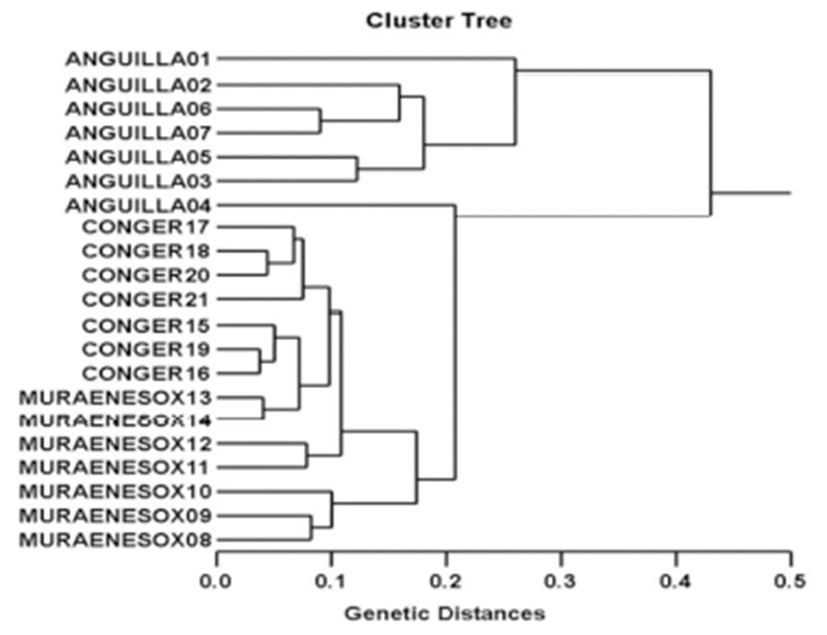
The seven oligonucleotides primers were shown to generate the shared loci, specific loci, unique shared loci to each species and shared loci by the three species which could be obviously scored. In the present study, 7 oligonucleotides primers generated 191 specific loci in the AJ species, 226 in the MC species and 181 in the CM species, respectively, as illustrated in Table 1. The primer BION-02 generated the most loci (a total of 83), with an average of 11.86 in the AJ species. The oligonucleotides primer BION-22 generated the least loci (a total of 21), with an average of 3.00 in the CM species, in comparison to the other primers used. The specific loci generated by oligonucleotides primers exhibited inter-individual-specific characteristics, thus revealing DNA polymorphisms. The gDNA isolated from three eel species were amplified by PCR. Here, the seven oligonucleotide primers were used to generate the unique shared loci to each species and shared loci by the three eel species, as illustrated in Table 2. With regard to average bandsharing value (BS) results, individuals from Conger myriaster species (0.808) exhibited higher bandsharing values than did individuals from Muraenesox cinereus species (0.729) (P<0.05), as displayed in Table 3. The dendrogram resulted from reliable seven oligonucleotides primers, indicating three genetic clusters composed of group I (ANGUILLA 01~ANGUILLA 07), group II (MURAENESOX 08~MURAENESOX 14) and group III (CONGER 15~ CONGER 21), as illustrated in Fig. 1. The longest genetic distance (0.430) displaying significant molecular difference was also between individual no. 01 within Anguilla japonica eel species and individual no. 04 within Anguilla japonica species. In the present study, PCR analysis revealed a significant genetic distance among three eel species. The existence of species differentiation and DNA polymorphisms among three eel species were detected by PCR analysis. This displays that the method is one of the appropriate tools for comparing the DNA of individuals, populations and/or species of eel. However, more researches will also be necessary to identify the discrimination between/among populations and species, using an annealing control primer system. From what has been said above, the potential of this analysis to ascertain diagnostic markers for the identification of three eel species has also been verified (Tasanakajon et al., 1998; Jing & Li, 1999; McCormack et al., 2000; Kimura et al., 2004; Kim et al., 2006; Yoon, 2008). It was reported that the species relation-ships revealed by the PCR approach should be consistent with previously obtained data using morphological portrait (Nebauer et al., 2000). As mentioned above, a dendrogram revealed close relationships between individual identities within three eel species. Thus, this PCR analysis revealed a significant genetic distance among the three eel species. High levels of a significant genetic distance among three eel species showed this PCR approach is one of the most suitable tools for individuals and/or species biological DNA studies. Therefore, this method can also be applied to other order of Anguilliformes and make technically-convenient the analysis of many samples in a short time.
Fig. 1.
Hierarchical dendrogram of genetic distances obtained from three eel species.
The relatedness among different individuals of Korean eel species from Anguilla japonica (ANGUILLA 01 ~ ANGUILLA 07), Muraenesox cinereus (MURAENESOX 08 ~ MURAENESOX 14) and Conger myriaster (CONGER 15 ~ CONGER 21) generated according to the bandsharing values and similarity matrix.
Download Original Figure
Table 1.
The number of total loci, average loci per lane, specific loci by PCR analysis using 7 oligonucleotides primers from three eel species in the Yellow Sea of Korea
| Item |
No. of average loci per lane |
No. of specific loci |
|
|
| Primer |
AJ |
MC |
CM |
AJ |
MC |
CM |
| BION-02 |
11.86 |
11.29 |
10.71 |
56 |
49 |
63 |
| (83) |
(79) |
(75) |
| BION-14 |
8.29 |
9.29 |
9.71 |
37 |
44 |
19 |
| (58) |
(65) |
(68) |
| BION-22 |
3.71 |
3.57 |
3 |
12 |
11 |
14 |
| (26) |
(25) |
(21) |
| BION-30 |
7.43 |
10.29 |
9.71 |
17 |
44 |
26 |
| (52) |
(72) |
(68) |
| BION-34 |
8.71 |
8.71 |
8.57 |
26 |
19 |
25 |
| (61) |
(61) |
(60) |
| BION-35 |
4.71 |
6.57 |
6.43 |
(33) |
(46) |
(45) |
| 33 |
39 |
24 |
| BION-40 |
4.43 |
4.86 |
4.43 |
10 |
20 |
10 |
| (31) |
(34) |
(31) |
| Total No. |
49.14 |
54.57 |
52.57 |
191 |
226 |
181 |
| (344) |
(382) |
(368) |
| Average no. per primer |
49.14 |
54.57 |
52.57 |
27.29 |
32.29 |
25.86 |
Download Excel Table
Table 2.
The number of unique shared loci to each species and number of shared loci by the three species generated by PCR analysis using 7 oligonucleotides primers in Korean eel fish such as Anguilla japonica (AJ), Muraenesox cinereus (MC) and Conger myriaster (CM), respectively
| Item |
No. of unique loci to each species |
No. of shared loci by the three species |
| Primer \ species |
AJ |
MC |
CM |
Three species (7 individuals per species) |
| BION - 02 |
27 |
30 |
12 |
126 |
| BION - 14 |
21 |
21 |
49 |
21 |
| BION - 22 |
14 |
14 |
49 |
21 |
| BION - 30 |
35 |
28 |
42 |
63 |
| BION - 34 |
35 |
42 |
35 |
28 |
| BION - 35 |
0 |
7 |
21 |
0 |
| BION - 40 |
21 |
14 |
21 |
42 |
| Total no. |
153 |
156 |
187 |
301 |
| Average no. per primer |
21.86 |
22.29 |
26.71 |
43 |
Download Excel Table
Table 3.
Multiple comparisons of average bandsharing values among three eel species were created according to the bandsharing values and similarity matrix.
| Species |
A. japonica
|
M. cinereus
|
C. myriaster
|
|
A. japonica
|
0.755±0.065b |
0.661±0.053a |
0.672±0.042a |
|
M. cinereus
|
- |
0.729±0.084b |
0.728±0.060b |
|
C. myriaster
|
- |
- |
0.808±0.037c |
Download Excel Table
ACKNOWLEDGEMENTS
Particular thanks go to the anonymous reviewers who aided them with thorough and far-reaching criticisms. The author thanks our laboratory colleague, Mr. J. H. Jeon and other undergraduate students for their support with sample collection, PCR procedures, and statistical analysis transactions.
REFERENCES
Jing QJ, Li YP. Random amplified polymorphic DNA analysis of eel genome. Cell Res. 1999; 9 p. 217-223.

Kim JR, Jung CH, Kim YH, Yoon JM. Genetic variations in geographic venus clam (Gomphina aequilatera, Sowerby) populations from Samcheok and Wonsan. Dev Reprod. 2006; 10 p. 227-238.

Kimura Y, Ishikawa S, Tokai T, Nishida M, Tsukamoto K. Early life history characteristics and genetic homogeneity of Conger myriaster leptocephali along the east coast of central Japan. Fish Res. 2004; 70 p. 61-69.

McCormack GC, Powell R, Keegan B. Comparative analysis of two populations of the brittle star Amphiura filiformis (Echinodermata: Ophiuroidae) with different life history strategies using RAPD markers. Mar Biotechnol. 2000; 2 p. 100-106.

Min MS, Yang SY. Isozyme and mtDNA analyses of the genus Anguilla (Pisces, Anguillidae). Korean J Zool. 1993; 36 p. 545-555.

Nebauer SG, del Castillo-Agudo L, Segura J. An assessment of genetic relationships within the genus Digitalis based on PCR-generated RAPD markers. Theor Appl Genet. 2000; 100 p. 1209-1216.

Oh TY, Jeong SB, Cho ES. DNA analysis of mtDNA COI gene in the sharp-toothed eel (Muraenesox cinereus Forskal) from Yeosu, Jinhae, Jeju, Goseoung, Jangheung and Haenam populations in Korea using PCR-aided RFLP. J Environ Sci. 2011; 20 p. 551-554.

Tassanakajon A, Pongsomboon S, Jarayabhand P, Klinbunga S, Boonsaeng V. Genetic structure in wild populations of black tiger shrimp (Penaeus monodon) using randomly amplified polymorphic DNA analysis. J Mar Biotech. 1998; 6 p. 249-254.

Yoon JM. Variability in two species of Osmeridae (Hypomesus nipponensis and Mallotus villosus). Dev Reprod. 2008; 12 p. 151-158.