INTRODUCTION
This scallop (Chlamys farreri) is, environmentally warmwater bivalve species, belonging to family Pectinidae, widely distributed on the coast of the Yellow Sea in the Korean Peninsula, as well as in several areas in Japan, South China Sea, Philippines, Indonesia under the natural surroundings. The clam is innate to benthic parts consisting of the rocks and pebbles regions from intertidal to shallow subtidal zone of 10 m in depth. Clams are also one of the most popular marine products in the Korean Peninsula because of their sense of taste and nourishing significance, and Koreans consume them abundantly. The coarse surface of this crustacean species is light gray and sculptures strong protuberance with scale-type, and the inside is white under the natural conditions. The shell is fan-type, and red-purple in color. Generally, there are marked differences of the shell weight, size, color and shape in Chlamys farreri according to the environmental circumstances of habitat such as prey, rock crystal, and water temperature. However, these kinds of Korean bivalve, which are recognized important physiologically (Oh et al., 2002; Park et al., 2012; Kim et al., 2015), reproductively (Chang et al., 1997; Park et al., 2006; Park & Lee, 2008), as well as histologically (Park et al., 2006), are not genetically and/or molecular-biologically studied like other crustaceans. Principally, there are marked differences of the size, color and shape in Chlamys farreri in keeping with the ecological conditions of habitat such as nourish and hard period. There is a need to understand the genetic traits and composition of this clam population in order to evaluate precisely the patent genetic consequence. PCR-based molecular techniques have been applied to examine the genetic characters of various life organisms. Above all, the polymorphic and/or specific markers specific to the line, the species, the genus or the geographical populations have been applied for the of individuals and species, hybrid parentage and for the screening of DNA markers (Partis & Wells, 1996; Callejas & Ochando, 1998; Muchmore et al., 1998; McCormack et al., 2000; Park et al., 2005; Song & Yoon, 2013; Oh & Yoon, 2014). So far, specific fragments generated by this PCR method using arbitrary primers had good advantages for detecting DNA similarity and diversity between living being (McCormack et al., 2000). The polymorphisms are determined by the banding patterns of amplified products at the specific positions by primers (Tassanakajon et al., 1998; Yoon & Kim, 2004). Above all, the clustering analysis of the genetic distance between genera/species/populations of various fishes and crustaceans from the different geographic locates has been performed using PCR research is of small number (Kim et al., 2000; Tassanakajon et al., 1998; Klinbunga et al., 2000; McCormack et al., 2000; Yoon & Park, 2002). The author undertook clustering analyses to disclose the Euclidean genetic distances between Korean and Chinese scallop population.
Materials and Methods
Adductor muscle tissues were collected separately from two Chlamys farreri populations of Korean scallop population (KSP) and Chinese scallop population (CSP), respectively. Genomic DNA samples isolated from KSP and CSP population were PCR-amplified repeatedly. PCR research was performed on DNA samples extracted from a total of 22 individuals using eight oligonucleotides primers.
DNA extraction should be accomplished according to the separation and extraction methods (Oh &Yoon, 2014). After several washings, lysis bufferⅠ(155 mM NH4Cl; 10 mM KHCO3; 1 mM EDTA) was added to the samples, and the mixture tubes were tenderly inverted. 600μL of chloroform was added to the mixture and then inverted (no phenol). Ice-cold 70% EtOH was added, and then the samples were centrifuged at 19,621 g for 5 minutes to extract the DNA from the lysates. The concentration of the extracted genomic DNA was measured with the optical density value at 260 nm by a spectrophotometer (Beckman Coulter, Buckinghamshire, UK). The DNA pellets were then incubation-dried for more than 12 hours, maintained at -40℃ until needed and then dissolved in the distilled water. Amplification products were separated by electrophoresis in 1.4% agarose gels with TBE, using DNA ladder (Operon Technologies, Alameda, CA, USA) as DNA molecular weight marker and detected by staining with ethidium bromide. The author used the oligonucleotides primers to certify the genetic distances of Chlamys farreri individuals. Eight oligonucleotides primers (Operon Technologies, Alameda, CA, USA), OPD-01 (5’-ACCG CGAAGG-3’), OPD-02 (5’-GGACCCAACC-3’), OPD-04 (5’-TCTGGTGAGG-3’), OPD-08 (5’-GTGTGCCCCA-3’), OPD-09 (5’-CTCTGGAGAC-3’), OPD-15 (5’-CATCCGT GCT-3’), OPD-18 (5’-GAGAGCCAAC-3’), and OPD-19 (5’-CTGGGGACTT-3’) were shown to generate the unique shared loci to each population and shared loci by the two scallop populations. Thus, the author used the primers to study the genetic variations and DNA polymorphisms of the scallop population. PCR was performed using programmable DNA Thermal Cycler (MJ Research Inc., Waltham, MA, USA) repeatedly. Eight oligonucleotides primers were shown to generate the unique shared loci to each population and shared loci by the two populations that could be obviously counted. After electrophoresis, gels were stained with ethidium bromide, illuminated with ultraviolet ray, and then photographed by photoman direct copy system (PECA Products, Beloit, WI, USA). Similarity matrix including bandsharing values (BS) between different individuals in the two scallop populations was produced according formula of Jeffreys & Morton (1987) and Yoke-Kqueen & Radu (2006). The average of within-population similarity was calculated by pairwise comparison between individuals within a population. A hierarchical clustering tree was assembled using similarity matrices to yield a dendrogram, which was assisted by the Systat version 10 (SPSS Inc., Chicago, IL, USA).
Results and Discussion
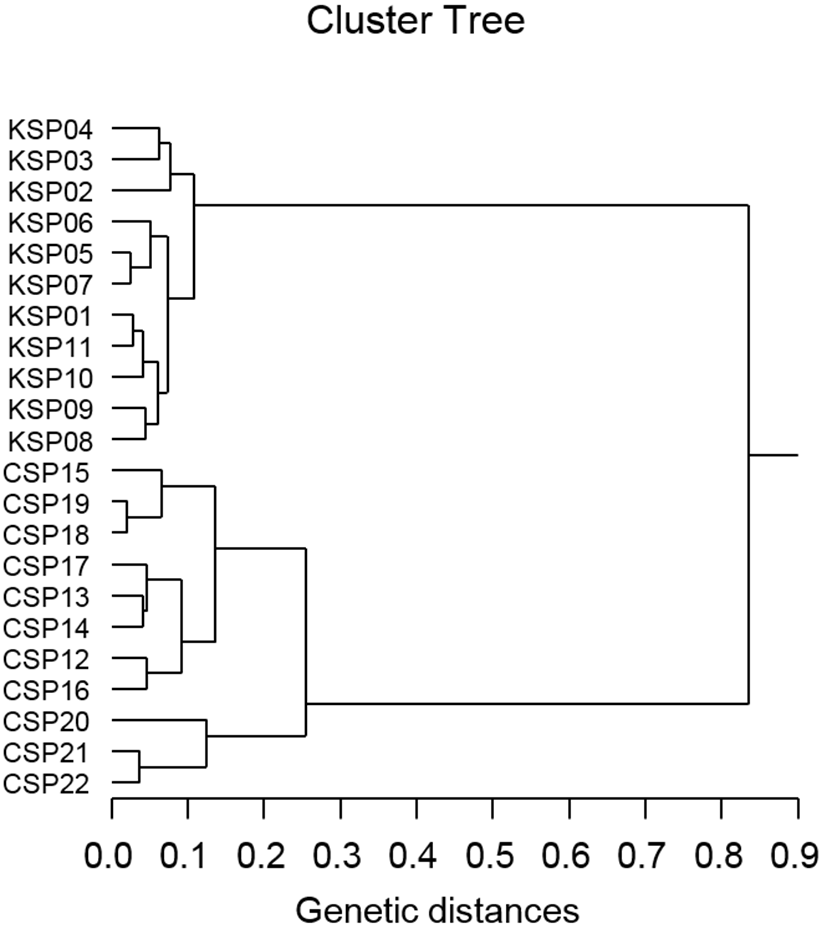
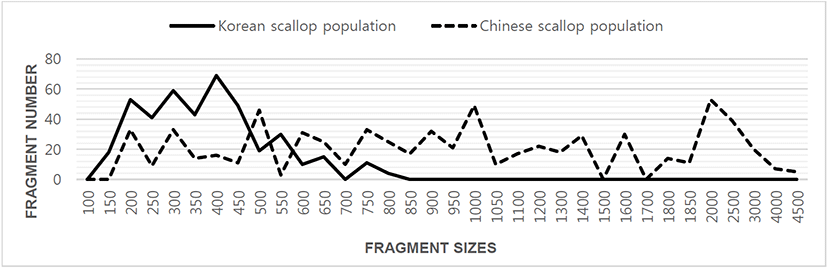
Author assessed genetic variation among individuals of the Korean scallop population and Chinese scallop population of Chlamys farreri, respectively, collected in the Yellow Sea and a site of Chinese sea by PCR analysis. DNA fragments acquired by eight oligonucleotides primers ranged in size from 100 to 4,500 bp in the scallop, as revealed in Fig. 1. The number of unique shared loci to each scallop population and number of shared loci by the two scallop populations generated by PCR research using eight oligonucleotides primers in the two scallop populations, respectively, as summarized in Table 1. The oligonucleotides primer OPD-01 generated 55 unique loci to each population, approximately 200, 250, 300, 400 and 450 bp, respectively, in the Korean scallop population. Specially, the primer OPD-09 generated 11 unique loci to each population, which were identifying each population, approximately 150 bp, in the Korean scallop population. The primer OPD-08 generated 77 unique loci to each population, which were determining each population, 500, 800, 1,000, 1,400, 2,000, 2,500 and 3,000 bp, in the Chinese scallop population. Remarkably, the primer OPD-15 distinguished 44 shared loci by the two scallop populations, major and/or minor fragments of sizes, which were identical in almost all of the samples. Numerous investigators researched the sizes of DNA fragments in the PCR structures of Eastern Pacific abalone (genus Haliotis) (Muchmore et al., 1998), blacklip abalone (Huang et al., 2000), Pollicipes mitella population (Song & Yoon, 2013) and mollusk species (Oh & Yoon, 2014). Multiple comparisons of average bandsharing values among scallop populations from two regions were generated according to the bandsharing values and similarity matrix, as illustrated in Table 2. Individuals from KSP population (0.798 ± 0.060) exhibited lower bandsharing values than did individuals from CSP population (0.832 ± 0.073) (P< 0.05). In the end, the average bandsharing value of this study is higher than that the between bullhead population (0.504 ± 0.115) (Yoon & Kim, 2004) and Spanish barbel species (0.71-0.81) (Callejas & Ochando, 1998). As displayed in Fig. 2, the dendrogram obtained by the eight oligonucleotides primers indicates two genetic groups: cluster 1 (KSP 01, 02, 03, 04, 05, 06, 07, 08, 09, 10 and 11) and cluster 2 (CSP 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 and 22). Among the twenty-two scallops, the shortest genetic distance that displayed significant molecular differences was between individuals 18 and 19 from the CSP population (genetic distance = 0.021), while the longest genetic distance among the twenty-two individuals that exhibited significant molecular differences was between individuals KSP no. 07 and CSP no. 15 (genetic distance = 0.835). The genetic distance between individuals approved the existence of close relatedness in the Korean scallop population. Comparatively, individuals of KSP population were fairly distantly related to that of CSP population, as shown in the hierarchical dendrogram of Euclidean genetic distances. The inter-population similarity indices and genetic distance values indicated that the Jamuna-Padma population pair of the Indian major carp (Catla catla) was genetically closer than the Halda-Jamuna and the Halda-Padma population pairs, which agreed with the geographical distances between them (Islam et al., 2005). They suggested that the RAPD technique could be used to discriminate between different river populations of major carp. A phylogenetic tree was constructed using UPGMA cluster analysis based on a total of 3,744 distinguishable fragments in gynogenetic clones from the silver crucian carp, Carassius auratus gibelio Block (Zhou et al., 2000). Clones A and P were the most closely related, whereas the most divergence was observed between clone D and clones E or F. The phylogenetic relationships among the 5 Haliotis species and one hybrid were carried out by calculating the distance coefficient and constructing a phylogenetic tree based on the PCR data (Kim et al., 2000). They were branched off into two clusters. Cluster I was formed by H. discus hannai, H. discus, H. gigantea, H. sieboldii, and the hybrid, which was subsequently divided into two sub-clusters. Cluster II contained only H. diversicolor aquatilis. Ultimately, they reported that PCR- bounded analysis is a powerful tool for determining the phylogenetic relationship between 6 Haliotis species. In invertebrates, cluster analysis of the pairwise population matrix, generated from genetic data, showed that geographically close populations be inclined to cluster together in the blacklip abalone (Huang et al., 2000). Overall, the population grouping of Chlamys farreri is established on morphological variations in shell weight, shell length, shell width, shell type, shell color and feet length. The prospective of oligonucleotides primer amplified polymorphic DNAs to determine diagnostic markers for breed, population, species and genus identification in shellfish has also been well identified (Partis & Wells, 1996; Tassanakajon et al., 1998; Huang et al., 2000; Kim et al., 2000; Klinbunga et al., 2000; McCormack et al., 2000; Park et al., 2005; Song & Yoon, 2013; Oh & Yoon, 2014). PCR fragments revealed in this study may be valuable as a DNA marker the two geographical populations to discriminate.
| Population | KSP | CSP |
|---|---|---|
| KSP | 0.798±0.060 b |
0.185±0.030 a |
| CSP | - | 0.832±0.073 c |