INTRODUCTION
Sea cucumbers are valuable marine resources supporting coastal livelihoods around the world. Of these, the sea cucumber, Apostichopus japonicus (Echinodermata: Holothuroidea), is one of the most economically important species inhabiting the shallow shelf of coastal regions around the Korean Peninsula, China, Japan (main island), and the Russian Far East (Liu, 2015). Color variation is an important trait in the sea cucumber aquaculture industry, which affects the market price of holothurian products (Jiang et al., 2013; Ru et al., 2019). Environmental factors, such as food and habitat, were found to determine the body color of sea cucumbers (MOF, 2004; Jiang et al., 2013). Interestingly, the red type sea cucumber showed definite genetic differentiation from the green and black types (Kan-No & Kijima, 2003). A report also suggested that the body color is affected by genetic factors in the parental generation (Park, 2006), partially supported by significantly different single-nucleotide polymorphism genotypes in allele-specific HSP70 gene between color variants (Kang et al., 2011). However, several studies have shown that there is no reproductive isolation and significant species-specific nucleotide polymorphism between color variants (Hu & Li, 2009; Jo et al., 2017). As the genetic distance between color variants (red, black, and green) of A. japonicus was 0.76%, irrespective of their body color, sea cucumbers can be classified as the same species (Sun et al., 2010). Recently, several transcriptomic and metabolomic analyses provided information for the potential molecular mechanisms underlying pigmentation, and color-determining genes and metabolites (Xing et al., 2017, 2018, 2021).
Light is an environmental factor that affects the feeding behavior, growth, and survival of aquatic animals directly or indirectly (Zhou et al., 1999). In addition, appropriate light intensities induced better growth and development of larval and juvenile sea cucumbers, and the photoperiod has diverse effects on sea cucumbers, for example, emerging at night and finding shelter during day-time (Zhang et al., 2006; Dong et al., 2010, 2011; Li et al., 2019). A comparison of juvenile sea cucumbers at different photoperiods showed the highest growth level at 12 h light and 12 h dark condition (Chen et al., 2007). However, further elucidation of the environmental and endogenous factors is required to fully understand the mechanism underlying pigmentation of juvenile sea cucumber. Recently, long-chain acyl-coenzyme A dehydrogenase (LCAD), nuclear distribution C-containing protein 3 (NUDCD3), and receptor tyrosine kinase Tie-1 (TIE1), whose expression levels increase at each pigmentation stage, were suggested to be involved in body color related proteins (Xing et al., 2017). In this study, we investigated the transcript levels of LCAD, NUDCD3, and TIE1 genes in swimming larva and juveniles of A. japonicus to obtain basic information for the relationship between expression of the body color related genes and pigmentation.
MATERIALS AND METHODS
In June 2021, the larva and juvenile sea cucumbers used in the experiments were produced by 4 h heat-shock stimulation (24°C from 20°C) of mature female and male sea cucumbers (n=200, mean body weight 200 g) that were separated by body colors (brown and black; Wondeok Fishermen’s Union, Samcheok, Korea). The fertilized eggs, developing embryos, larva, and juveniles were maintained at 21°C with filtered seawater and air supplied using an air blower. Commercial feed of spirulina powder (Shandong, China) and bakers’ yeast (AngelYeast, Hubei, China) was supplied once a day to the swimming larva (until 20 days post-fertilization, dpf) and increased to twice a day (90-dpf) after the juveniles attached. Seawater was changed twice a day using a siphon and overflow during the experimental period.
To examine effect of light on the body color, the larval sea cucumbers were divided into two groups (LED-exposed/ light-shielded) and further reared at 200 L plastic tanks from 30-dpf to 90-dpf. One group was exposed to LED lights (3,000 lx; MB-600S, Mobydick, China) for 10 h in a day (LED-exposed condition, light phase (L): dark phase (D)=10/14 h), and the other group was covered with a PVC board during the experimental period, except for feeding time (light-shielded condition, L:D=0/24 h) (Table 1). The light exposure time was based on the average sunrise/sunset time at Samcheok, Gangwon-do, Korea, June 2021. This study did not require approval by the institution and ethics committee because there are no vertebrate participants.
dpf, days post-fertilization; NA, not applicable.
Nucleotide sequences for LCAD, NUDCD3, and TIE1 were retrieved using the Chinese Sea Cucumber Database (http://www.genedatabase.cn/aja_genome_20161129.html). According to the sequence information in the database, the primers were designed using the ApE program (A plasmid Editor Version 2.0.36. by M. Wayne Davis) and synthesized by Bioneer (Daejeon, Korea; Table 2). Total RNAs were extracted from the intestines and the respiratory trees of mature sea cucumbers as previously described (Maeng et al., 2019). The LCAD, NUDCD3, and TIE1 cDNAs were amplified by PCR methods using primer sets (Table 2) and LA Taq (Takara, Osaka, Japan) and inserted into pGEM-T Easy vector (Promega, Madison, WI, USA). The PCR cycling conditions for LCAD and TIE1 were as follows: 1 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 55°C, and 35 s at 72°C; 10 min at 72°C. For NUDCD3, the PCR conditions were as follows: 1 min at 94°C; 32 cycles of 10 s at 98°C and 30 s at 68°C; 10 min at 72°C. PCR products were purified using PCR and/or Gel Purification Kit (Bioneer) and ligated with pGEM-T Easy vector for 24 h at 16°C using T4 DNA ligase (Promega). The pGEM-T Easy vectors containing LCAD, NUDCD3, and TIE1 cDNAs were transformed into DH5 α competent cells and cultured in Luria-Bertani ampicillin medium for 24 h at 37°C. The pGEM-T easy vectors were extracted using a plasmid mini extraction kit (Qiagen, Valencia, CA, USA) and the inserted cDNAs were sequenced by Sanger method (Solgent, Daejeon, Korea). The nucleotide sequences were analyzed using the MultAlin multiple sequence alignment site (http://multalin.toulouse.inra.fr/multalin/) and the ApE program.
LCAD, long-chain acyl-coenzyme A dehydrogenase; NUDCD3, nuclear distribution C-containing protein 3; TIE1, tyrosine kinase Tie-1.
Total RNAs were extracted from juvenile sea cucumbers sampled at 40-, 50-, and 60-dpf (10, 20, and 30 days after LED exposure) using the RNeasy Mini Kit (Invitrogen, Waltham, MA, USA). To synthesize cDNAs, PrimeScript RT reagent Kit (Takara) was used along with total RNA (1 μg each), 5× gDNA Eraser Buffer, 2 μL of gDNA Eraser (1 μL), and the PCR mixtures including 5× PrimeScript Buffer II, 4 μL PrimeScript RT Enzyme Mix I, 1 μL RT Primer Mix, 1 μL RNase-Free distilled water were reacted for 2 min at 42°C. Reverse transcription was performed for 15 min at 37°C and incubated at 85°C for 5 s to inactivate reverse transcriptase to synthesize cDNAs. qPCR was performed using the QuantStudio™ 7 Flex Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). The primers used in qPCR were designed using Primer Express v3.0 software (Applied Biosystems) and synthesized by Bioneer (Table 2). Using 10 ng of the synthesized cDNA as a template, primers (10 μ M), TB GreenTM Premix EX Taq II (Takara), and ROX Reference Dye II (Takara) were used, and the final qPCR mixture was a two-step qPCR with a volume of 20 μL. qPCR conditions were 95°C for 30 s; 40 cycles of 95°C for 5 s and 60°C for 34 s; 95°C for 15 min and 60°C for 1 min.
Larval specimens (1.5–9 dpf) were hybridized in situ using a digoxigenin-labeled LCAD and NUDCD3 antisense probes. Detection of mRNA was carried out essentially according to the protocol of Arenas-Mena et al. (2000) and Maeng et al. (2012).
Statistical significance was evaluated by one-way ANOVA using SPSS software (version 23.0; IBM, Armonk, NY, USA) and Tukey’s post hoc test. In addition, the Student’s t tests were used to compare the expression levels data between the LED-exposed and light-shielded conditions. Differences between the groups were considered significant at p<0.05.
RESULTS
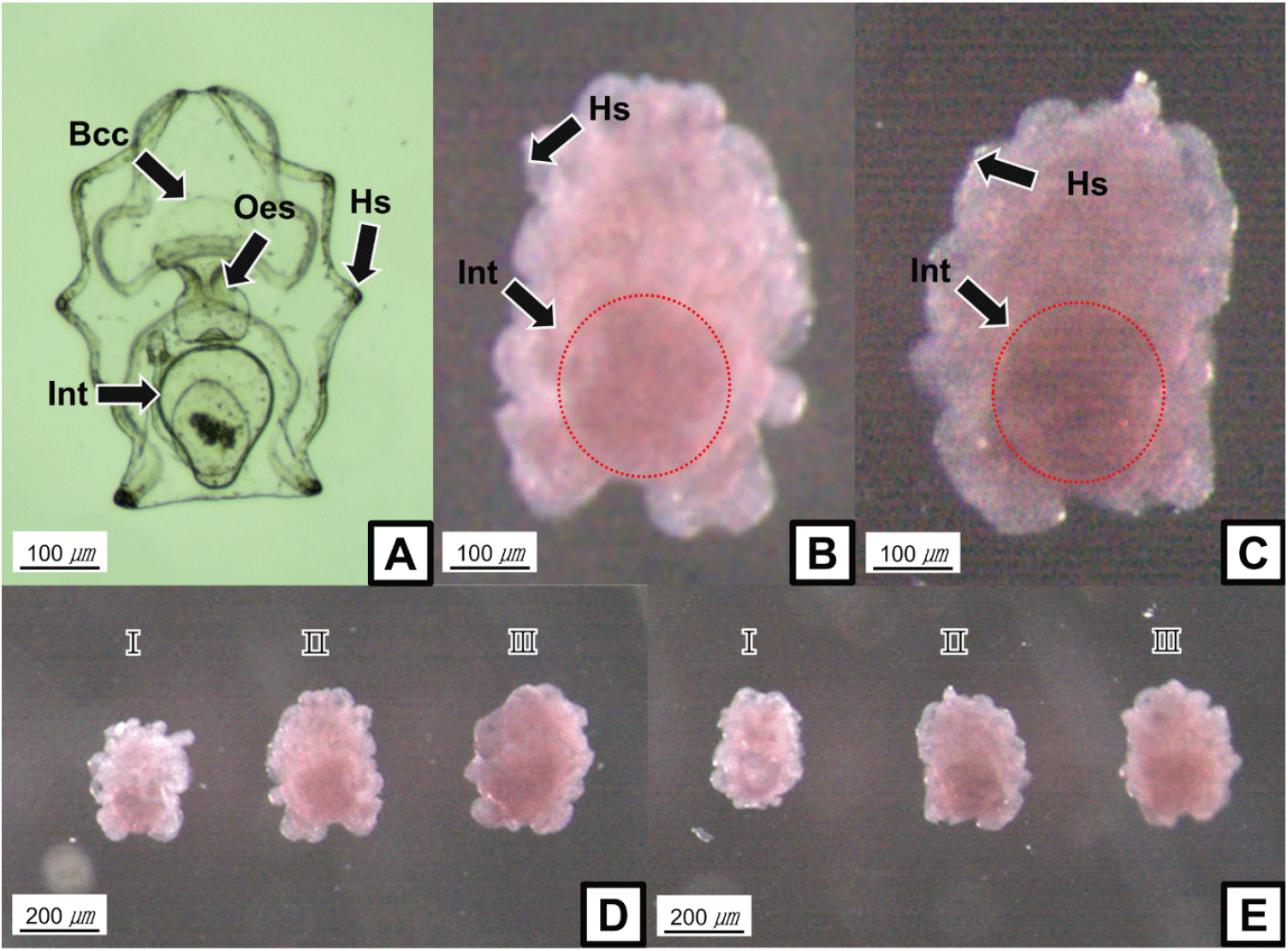
WMISH shows that sea cucumber LCAD and NUDCD3 transcripts are mainly localized to a population of cells in the internal organs of the auricularia larva, such as intestine, although the mRNAs could not be observed clearly (Fig. 1B and C). In addition, the gene expression levels showed increasing tendencies during each development stages of the auricularia larva (Fig. 1D and E).
In general, the examined LCAD, NUDCD3, and TIE1 transcript levels showed increasing tendencies during larval development stages (6-dpf to 60-dpf) of sea cucumbers. In light-shielded individuals, however, the LCAD, NUDCD3, and TIE1 mRNA were not significantly changed during 40-dpf and 60-dpf stages. At 60-dpf juveniles, the LCAD and TIE1 mRNA levels of LED-exposed individuals were significantly higher than those of light-shielded individuals (p<0.05; Fig. 2A and C). At 40-dpf and 50-dpf juveniles, however, the NUDCD3 mRNA expression levels were significantly higher in light-shielded condition than LED-exposed condition (p<0.05; Fig. 2B).
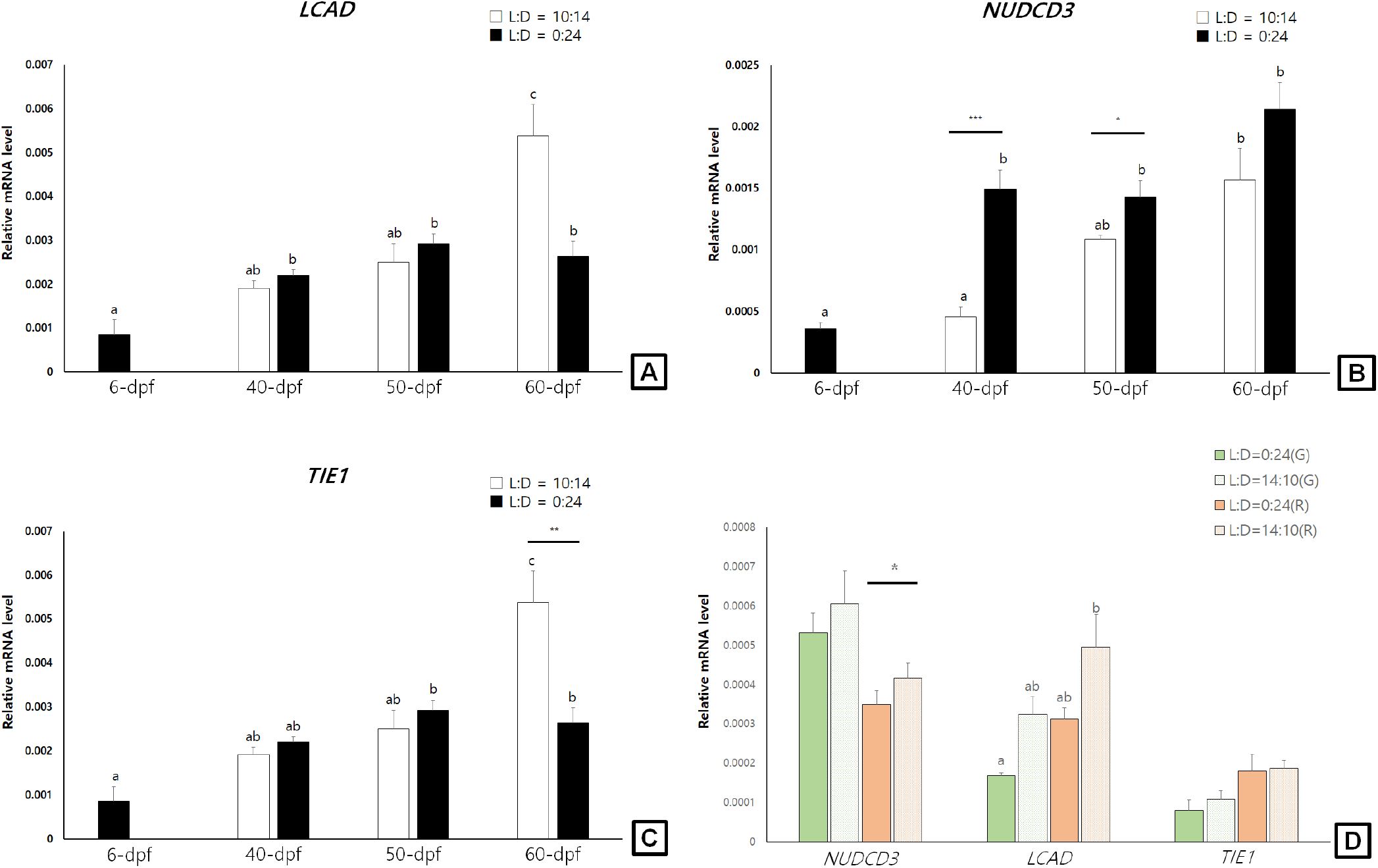
In the pigmented juveniles (90-dpf), the LCAD and TIE1 mRNA levels showed higher tendencies in the red individuals than those in green ones, whereas the NUDCD3 mRNA was higher level in the green juveniles (Fig. 2D). In addition, the LCAD mRNA levels were significantly higher in the LED-exposed red individuals compared to the light-shielded ones and the NUDCD3 mRNA level was slightly but significantly higher in the LED-exposed red juveniles (p<0.05).
DISCUSSION
More than 50 years ago, the body color of sea cucumbers was known to be determined by pigment cells in the dermis layer of the body wall in granule forms (Menton & Eisen, 1970). Among the different color morphs, red, black, and albino are rare and have great appeal to consumers as high-quality traits (Ru et al., 2019). In this study, we examined spatiotemporal expression profiles of the body color related mRNAs, LCAD, NUDCD3, and TIE1 at several developmental and pigmented stages of A. japonicus. In addition, LED-exposed and light-shielded juveniles were applied to the qPCR analysis to better understand the relationships among body color, LED-exposure, and the gene expression levels LCAD, NUDCD3, and TIE1.
In various mammalian melanocytes, tyrosine kinase receptors (RTKs), such as growth factor receptors and TIEs, produce a variety of signals associated with stimulation of proliferation, differentiation, migration, and survival (Zhou et al., 2019). In fact, activation of RTKs by a ligand induced transcription of tyrosinase and melanin synthesis in differentiated melanocytes, and expression of a mutant RTK was associated with lesser pigmentation of melanocytes, suggesting the downregulation of tyrosinases (Luo et al., 1995; Alexeev & Yoon, 2006). In accordance with the mammalian RTKs, we showed that the sea cucumber TIE1 transcript levels were gradually increased at juvenile stage nearby pigmentation period compared to those of the earlier developmental stages, irrespective of LED-exposed condition. Interestingly, the TIE1 gene expression level of LED-exposed sea cucumber juveniles was significantly higher than that of light-shielded juveniles, implying the possible involvement of light in TIE1-meditated pigmentation process.
Melanin is considered as the major pigment widely present in diverse animal phyla, and the variations in melanin formation result in a wide range of body colors including echinoderms (Calestani & Wessel, 2018). In the sea cucumber body, the concentration of melanin pigment was gradually increased with deepening color (Xing et al., 2021). In this study, LCAD expression increased at juvenile stage nearby pigmentation period, suggesting the potential role of LCAD in the synthesis and accumulation of melanin pigments. In 60-dpf juveniles, LED-exposure increased transcript level of LCAD than that of light-shielded sea cucumber, which are similar results for both protein and mRNA levels of LCAD during development of purple sea cucumber (Xing et al., 2017) and upregulation of LCAD protein level in human keratinocytes under radiation condition (Amar et al., 2019). In future studies, fine-tuned examination for diverse light sources and photoperiods are required to better understand the relationship between higher expression level of LCAD and body color of sea cucumber.
In differentially expressed proteins during pigmentation stages of purple sea cucumber, NUDCD3 was classified in two functional groups of melanogenesis and Wnt signaling pathway (Xing et al., 2017). Activation of the Wnt signaling pathway enhanced melanocyte specification at early stages of zebrafish development (Vibert et al., 2017), suggesting a pivotal role of Wnt signaling in regulation of melanocyte differentiation in vertebrates. Interestingly, we observed an increasing pattern of NUDCD3 expression levels in light-shielded 40- and 50-dpf juveniles compared to those of LED-exposed ones, whereas LED-exposure increased the NUDCD3 expression levels at later pigmentation stages. Considering the promoting effect of Wnt signaling in melanocyte development and pigmentation (Schepsky et al., 2006; Wang et al., 2017), it is noteworthy that the Wnt signaling pathway associated with melanin synthesis and regulation in sea cucumbers are likely dependent to light-exposure and developmental stages.
Spatial distribution of LCAD and NUDCD3 transcripts in swimming larva stage was expected to be high in the body surface. Unexpectedly, LCAD and NUDCD3 were shown to be highly expressed in internal organs such as intestine. The spatial gene expression related to the skeletonic mesenchyme lineage in sea cucumber larvae also showed similar patterns with the ingression of skeletogenic pigment cells in sea urchin embryos (McCauley et al., 2012). Thus, it is presumed that genes distributed in internal organs through various metamorphic processes, such as impregnation and rotation, migrate to the body surface and participate in the formation of the skeleton and body color. However, the detailed involvement of LCAD and NUDCD3 genes in pigmentation process of sea cucumbers remains to be further studied. To select and produce high-quality sea cucumbers, further studies are essentially required, such as evaluation of the body color related genes and characterization of pigmentation process in the melanocytes.







