INTRODUCTION
LHX8 is a member of the LIM homeobox gene family (Kitanaka et al., 1998). In mice, Lhx8 preferentially expressed in oocytes of germ cell clusters and primordial, primary, and antral follicles in the ovary (Choi et al., 2008; Pangas et al., 2006). Lhx8 deficient female mice are infertile, wherease knockout male is fertile. Lhx8 is required for development and differentiation of the oocytes during folliculogenesis after birth in the ovary (Choi et al., 2008). At the time of birth, histological structures of Lhx8 deficient (Lhx8–/–) ovaries are grossly similar to the wild-type ovaries (Choi et al., 2008). The number and morphology of primordial follicles in Lhx8 deficient ovaries shows no significant differences (Choi et al., 2008). However, the activation and differentiation of primordial follicles in Lhx8 deficient ovaries are arrested at primary follicle stage, and then they loses the follicles within postnatal day 7 (Choi et al., 2008). Based on the analysis of gene profiles from newborn ovaries of wild type and Lhx8 deficient mice, numerous oocyte-specific genes including such as zona pellucid genes 1~3 (Zp1~3), growth differentiation factor 9 (Gdf9), bone morphogenetic protein 15 (Bmp15), and Pou5f1 (also known as Oct4), Fig.la (Factor in the germline alpha), Nobox (Newborn ovary homeobox), Sohlh1 (Spermatogenesis and oogenesis specific basic helix-loop-helix transcription factor 1), and Pad6 (Peptidylarginine deiminases), are mis-regulated in Lhx8 deficient ovaries (Choi et al., 2008). However, it is unknown whether Lhx8 directly or indirectly regulates transcription of these oocyte-specific genes during development of the ovarian follicle. In the present study, we identified DNA sequences that Lhx8 binds and show that Lhx8 transactivates luciferase reporter containing Lhx8 DNA binding element (LBE).
METERIALS AND METHODS
To create the pET41b-Lhx8 homeodomain protein (GST-Lhx8HD) and bacterial expression construct, the insert was amplified by PCR and cloned into pET41b (Novagen, Madison, WI). We verified the sequences of the resulting GST-Lhx8HD fusion constructs to ensure that no mutations have been introduced during PCR cloning. We transformed BL21-pLysS Escherichia coli (Stratagene) with GST-Lhx8HD construct and expressed it by inoculating Luria-Bertani (LB) media containing 20 μg/ml kanamycin with overnight culture (1:20 dilution) at 37°C to an OD600 of 0.5. Protein expression was induced at 30°C with 2mM of isopropyl-1-thio-β-Dgalactopyranoside (IPTG) to OD600 of 1.0. We stored cell pellets at –80°C, thawed and lysed them with BugBuster lysis buffer (Novagen). We purified the GST-Lhx8HD fusion proteins with GST bind resin (Novagen), were dialyzed them three times in 1 liter of phosphate-buffered saline and quantified them. Purified proteins were stored at –80°C until use.
The pGL4-promoter vector with SV40 promoter (Promega) was used for constructing luciferase reporter vectors containing three repeats of LBE (3xLBE-Luc). Overexpression vector carrying the mouse Lhx8 were constructed by cloning the full-length of Lhx8 cDNA into pCMV-Tag3 or Tag5 vector (Agilent Technologies).
EMSA were performed in 20 μℓ reaction mixtures at room temperature at a final concentration of 10mM Tris, pH 7.5, 50 mM NaCl, 1.5 mM MgCl2, 2.5 mM dithiothreitol, 5% glycerol, 5 μg/ml poly(dI)-poly(dC), and 250 μg/ml bovine serum albumin. EMSA probes were prepared by end-filling annealed primers with [α-32P] dCTP and Klenow polymerase (Invitrogen). Binding reactions were conducted by incubating 32P-labeled probes (250,000 cpm/reaction) with 50 ng of purified proteins or 1 μg of ovarian extract in the absence and presence of polyclonal anti-GST (Amersham Biosciences). For competition assay, purified proteins were incubated at room temperature with cold competitors for 15 min before addition of the 32P-labeled probe.
The CAST assay was carried out essentially as previously described with some modifications (Choi et al., 2006). We used purified GST-Lhx8HD proteins and GST-bind resin (Novagen) for the CAST assay. Binding reactions for CAST were performed in DNA binding buffer (10 mM Tris. pH 7.5, 50 mM NaCl, 7.5 mM MgCl2, 1mM EDTA, 5% glycerol, 5% sucrose, 0.1% Nonidet P-40, and 5 mg/ml bovine serum albumin). 0.5 pmol of short double-stranded DNA containing a 15-bp random sequence flanked by 20-bp fixed sequences, CAST oligonucleotides was incubated with 100ng of purified GST-Lhx8HD protein for 20min at room temperature. Unbound DNAs were washed away with binding reaction buffer. Bound DNAs were amplified by PCR for the next round of CAST. A total of five cycles of CAST were performed. Final PCR products were cloned into the pGEM-T easy vector (Promega). The inserted DNAs were sequenced and scored to get the consensus sequences.
Human embryonic kidney cells (HEK293) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. For transient transfection, FuGENE6 (Roche Applied Science) was used according to the manufacturer's instructions. Following the transfection, the cells were incubated for 48 hr before harvest. For each transfection, 200 ng of reporter construct, 200 ng of the indicated expression plasmid, and 10 ng of pRT-TK normalization plasmid were used per single well of a 6-well plate. Dual luciferase assays were carried out with total cell extracts as recommended by Promega. The transient transfection experiments were performed in triplicate, and results were normalized to the expression of Renilla luciferase.
RESULTS
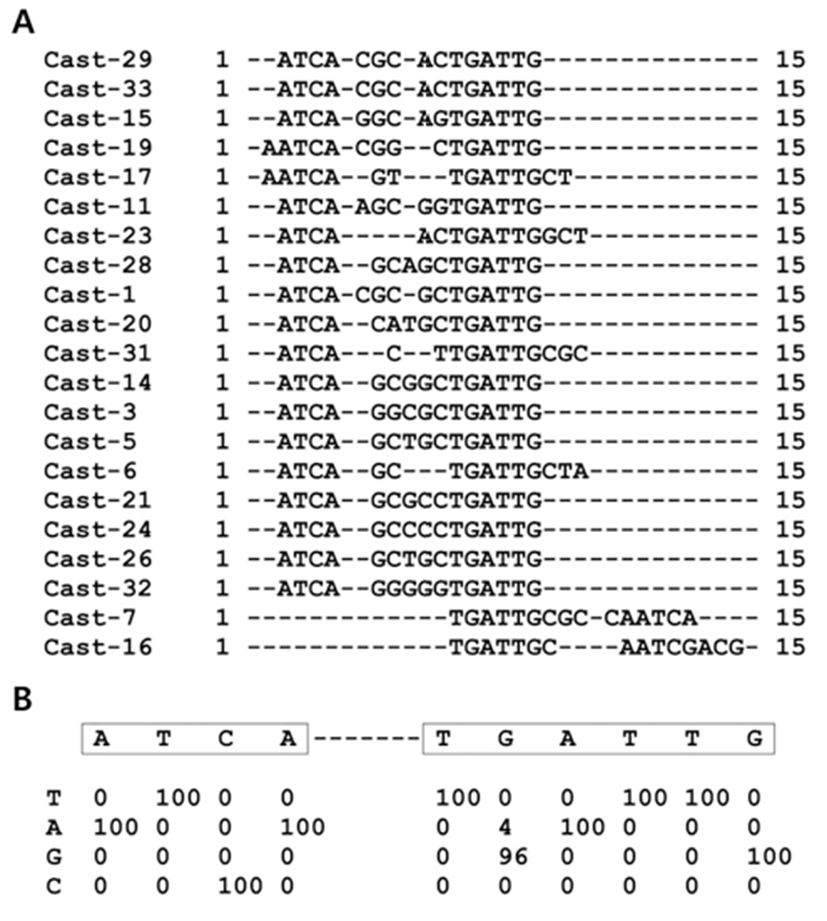
We determined a consensus DNA binding sequence for Lhx8 using the CAST assay as previously described (Choi et al., 2006). We used a predicted homeodomain portion of Lhx8 (a.a 246-308) fused to GST-Lhx8HD. DNA sequences that bound GST-Lhx8HD went through five cycles of CAST. The 21 selected sequences for GST-Lhx8HD were aligned (Fig. 1A) and scored (Fig. 1B). The most frequently observed sequences are shown in Fig. 1B based on the percentage occurrence of each base at each position from 96 to 100%. CAST assay revealed two groups of consensus sequence, ATCA-TGATTG (Fig. 1B), as the Lhx8 DNA Binding Element (LBE).
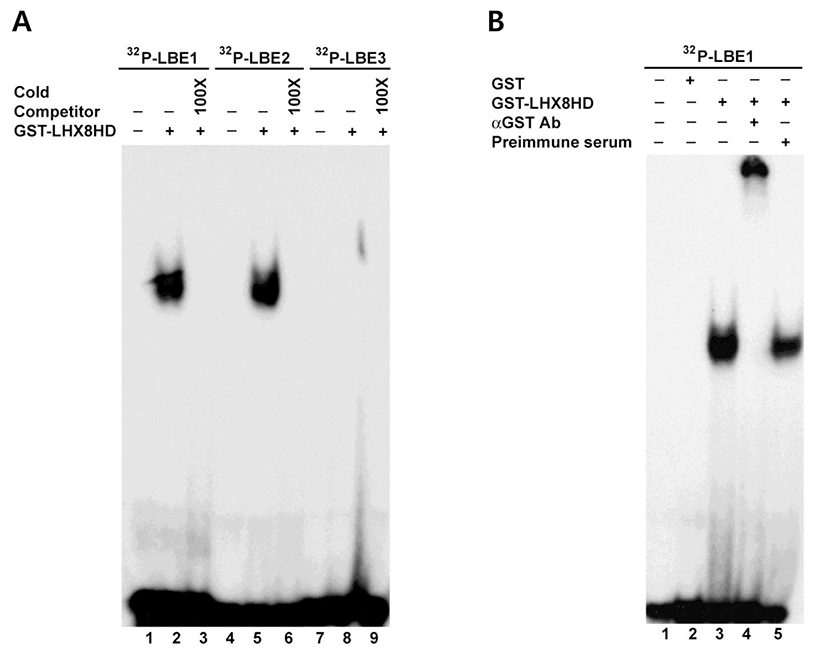
We next confirmed the interaction between LBE and GST-Lhx8HD using EMSA. Using a competitive binding assay (Fig. 2A), GST-Lhx8HD protein bound to 32Plabeled LBE1 containing sequences, TGATTG, or LBE2 (ATCATGATTG) probes (Fig. 2A). However, GSTLhx8HD protein did not form DNA-protein complex with 32P-labeled LBE3 (ATCA) probe (Fig. 2A). The bound complex was competed out by 100-fold molar excess of unlabeled cold each competitor (Fig. 2A). To further confirm the specificity of these DNA-protein complexes, anti-GST antibodies were added to the binding reaction in the absence or presence of GST-Lhx8HD or GST protein. As shown in Fig. 2B, the anti-GST antibody further shifted the DNA-protein complex (Fig. 2B). To confirm the effect of GST, preimmune serum was added to the binding reaction in the presence of GST-Lhx8HD (Fig. 2B).
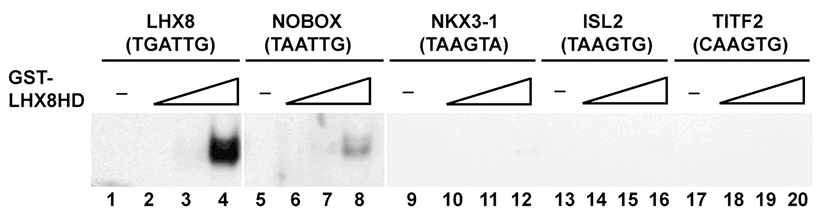
We examined whether the homeobox domain of Lhx8 has binding specificity with other homeobox consensus core sequences. Increasing amounts of purified GST-Lhx8HD protein incubated with 32P-labeled probes containing known other homeobox consensus core sequences, TAATTG for Nobox, TAAGTA for Nkx3.1, TAAGTG for Isl2, or CAAGTG for Titf2 were analyzed using EMSA. At high concentration (100 nanogram), GST-Lhx8HD protein bound to these related sequences with low affinity compared with the LBE (Fig. 3). Only the sequence, TAATTG showed low affinity with GST-Lhx8HD under high concentration in lane 8 (Fig. 3), whereas Lhx8 didn’t form DNA-protein complex with other related sequences. This result suggests that Lhx8 preferentially binds to the LBE containing 6 core sequences, TGATTG.
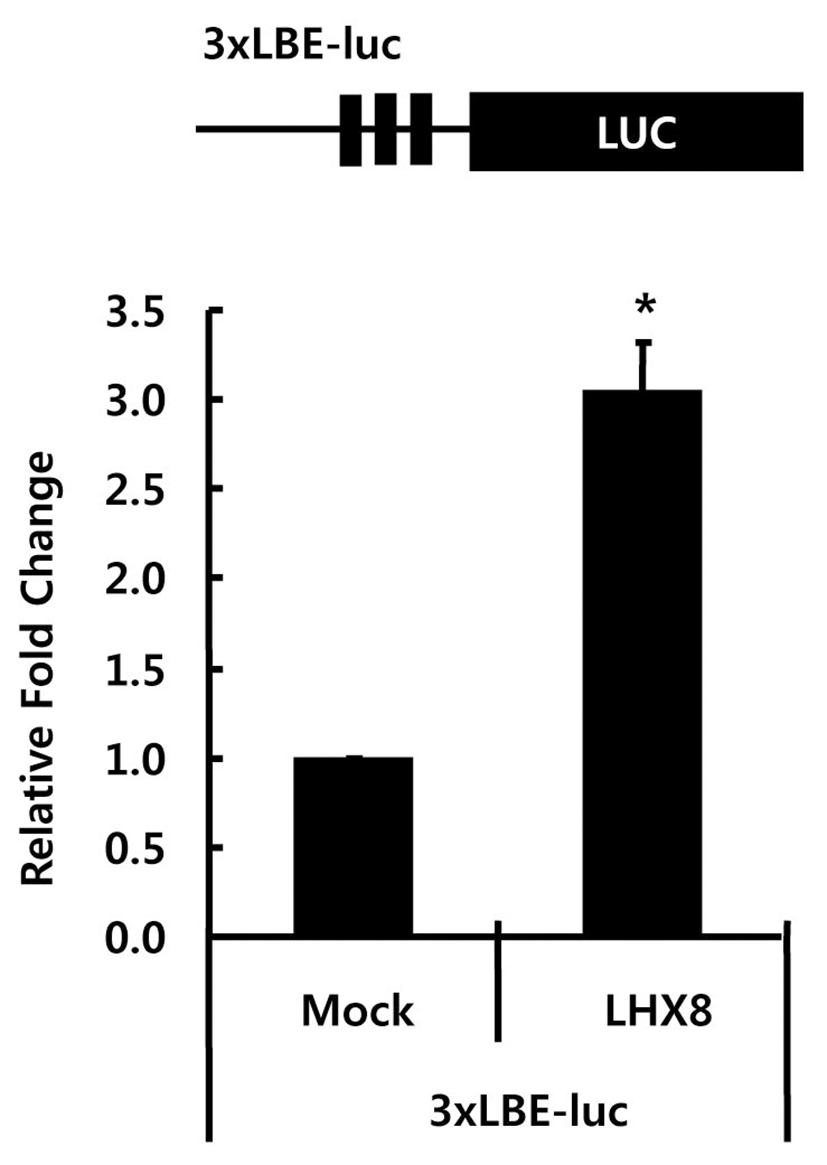
To examine whether Lhx8 can regulate the expression of LBE driven reporter gene in vivo, we constructed a luciferase reporter vector containing three LBEs (TGATTG), 3xLBE-pGL4 upstream of the luciferase reporter gene. This reporter vector was co-transfected with either empty vector (pCMV-Tag5) or Lhx8 over-expression vector (pCMV-Tag5) into HEK293 cells. The relative luciferase activity of reporter construct containing three copies of TGATTG was increased by 2.3-fold with the overexpression of Lhx8 (Fig. 4). This suggests that Lhx8 can activate the transcription of target genes through the LBE.
DISSCUSSION
In the previous study, we showed that numerous oocyte-specific genes, including Gdf9 and Nobox, were down-regulated in ovaries that lack Lhx8 (Choi et al., 2008). However, it is unknown whether Lhx8 directly or indirectly regulates transcription of these oocytespecific genes during development of the ovarian follicle. To address the direct downstream genes of Lhx8, it is required for characterization of Lhx8 as a transcription factor. In the present study, we have identified a consensus DNA binding sequence, TGATTG, as Lhx8 DNA binding elements using purified proteins in a cyclic amplification target assay (Fig. 1). The consensus DNA binding sequence of Lhx8 consists of six base pairs as core sequences of known homeoproteins including Msx1, Titf2, Nkx3-1, Isl2 and Nobox (Choi et al., 2006; Catron et al., 1993; Guazzi et al., 1990; Kim et al., 2003). The LBE core sequence for Lhx8 is slightly different from others; CAAGTG for Titf2, TAAGTA for Nkx3-1, TAAGTG for Isl2 and TAATTG/A for Nobox. This suggests that their functions might be redundant in the regulation of their down-stream targets. However, Msx1, Titf2, Nkx3-1,and Isl2 except Nobox, are not expressed in the newborn ovaries (data not shown). In the previous study, Nobox is exclusively expressed in the oocyte like Lhx8 (Rajkovic et al., 2004; Suzumori et al., 2002). We characterized a binding site and target genes of Nobox in the ovary (Choi et al., 2006; Choi et al., 2007). The Lhx8 DNA binding element, TGATTG, is similar to TAATTG/A, that of Nobox. This suggests that their functions might be overlapped in the regulation of their down-stream targets. However, each knockout mice show different phenotype (Choi et al., 2008; Rajkovic et al., 2004). Lhx8 deficiency looks more severe than Nobox deficiency. It suggests that the regulatory network of Lhx8 might be independent on that of Nobox. These results spotlight that the transcriptional activity of Lhx8 requires direct DNA binding through specific consensus sequences, LBE.