INTRODUCTION
Recently, land animals reach saturation as the breeding and emphasized the importance of fish as protein sources. Although a catch of fish is increased through successful development of fishing technology and aquaculture, aging of fisheries, recessive of breeding species, breeding density and occurrence of refractory disease lead to increase occurrence frequency of disease and massive mortality. Fish are free-living organisms from the embryonic stage of life in their aquatic environment, this environment is ideal for pathogen attraction (Gomez et al., 2013). Accordingly, protection mechanism is important beginning at the early stages of embryogenesis in fish.
The immune system is biological structures and processes within an organism that protects against disease. To function properly, an immune system must detect a wide variety of agents and distinguish self or non-self. Most pathogens altered host cells and can be recognized by immune cells through expressed pathogen- or danger-associated molecular patterns (PAMPS or DAMPS, respectively). But pathogens can rapidly evolve and adapt, and thereby avoid detection and neutralization by the immune system. To remove the invaders directly or destroy their ability to replicate, specialized immune cells of the innate and adaptive responses appeared during evolution (Beck & Habicht, 1996; Fischer et al., 2013).
The immune system of fish is similar to higher vertebrates, despite having some of the differences. In contrast to higher vertebrates, fish live in aquatic environments from early embryonic stages of life, which are an ideal medium for microorganism growth compared to air. Therefore, fish depend on their innate immune systems for survival. The innate immunity is a fundamental defense mechanism in fish even though a simple and straight-forward (Uribe et al., 2011; Bowden et al., 2005) whereas adaptive immune response is highly complex and specific.
The adaptive immune system is composed of a highly specialized, systemic cell which processes to eliminate or prevent pathogenic challenges. Adaptive immunity plays a vital role in protection against recurrent infections by generating memory cells (cell-mediated immunity) and specific soluble and membrane-bound receptors (humoral immunity). These receptor proteins identify molecular patterns that are typical of pathogenic microorganisms, including polysaccharides, lipopolysaccharide (LPS), pepti-doglycan bacterial DNA, viral RNA and other molecules that are not normally on the surface of multicellular organisms. A fundamental component of the adaptive immune system is the T cell which allow the fast and efficient elimination of the specific fish pathogens (Ellis, 2001; Swain, 2006).
Fish possess lymphocyte populations that are analogous to T cells, B cells, cytotoxic cells (similar to natural killer cells), macrophages and polymorphonuclear leukocytes. The immune system of teleosts has sub-populations of T lymphocytes that exhibit differential responses to mitogens, B cell acute allograft reactions, mixed leukocyte reactions and cooperative interactions between T cells, B cells and macrophages that are essential for the production of antibodies. Moreover, elasmobranch and teleost fish are the most primitive groups that possess the Major Histocompatibility Complex (MHC) and T cell receptor (TCR) (Manning & Nakanishi, 1996; Davis & Bjorkman, 1988; Miller and Withler, 1998).
The T cell has a membrane-bound receptor, which is responsible for antigen recognition and is composed of four distinct polypeptide chains (alpha, beta, gamma, and delta). There are two types of T cell populations based upon their receptor heterodimers (alpha/beta and gamma/delta). The polypeptide chains are derived from variable (V), joining (J), diversity (D), and constant (C) gene segments. These segments are randomly selected from the germ-line gene pool by a recombination mechanism to generate a wide diversity for antigen recognition (Davis, 1990).
The information on alpha/beta TCR is extensive that Ag-recognition processes, structure, and genetics due to alpha/beta T cells comprise ~90% of PBLs of humans (Haas et al., 1993; Kronenberg, 1994). The second class of T cells, termed delta T cells, is less well understood, as only 1–10% of human T cells express gamma/delta TCR. Nonetheless, the gamma/delta T cells may comprise one-half of the T cell population in various human tissues (gut, reproductive, and mucosal epithelia) or in the peripheral blood of ungulates. Thus, gamma/delta T cells are expected to have a fundamentally different role from alpha/beta T cells (Chien et al., 1996; Hayday, 2000).
In this study, we observed the temporal expression of olive flounder TCR subunit genes in the early stage and analyzed expression in the organ in adult fish. This will provide key information for the understanding of fish immune system at early stage of development.
MATERIALS AND METHODS
Olive flounders were obtained from Genetics and Breeding Research Center, National Fisheries Research and Development Institute (NFRDI; Geoje, Republic of Korea), and maintained in 10 tons flow through tank at 20±1°C under a natural photoperiod. The sample was prepared from various tissues including brain, muscle, fin, eye, liver, spleen, kidney, and gill obtained from 2-year-old healthy olive flounder (38 cm±SEM, n=3). Different stages of embryo (0.92±0.02 mm), larvae (2.49±0.34 mm) and juvenile (3.5±0.45 mm) development were described from fish kept at 20°C in the tank. The sample of 10 randomly selected embryo or fish were collected including fertilized egg (F), blastula (B), neurula (N) including 0 to 35 days post hatching (dph) (H0, H1, H3, H7, and H35) and immediately frozen in liquid nitrogen, and stored at –80°C until RNA extraction.
The amino acid sequences of the four TCR reported from GenBank, accession numbers are as follows: PoTCR-alpha (BAB61895), PoTCR-beta (BAB61899), PoTCR-gamma (BAC00869), PoTCR-delta (BAC00868) and aligned using the GeneDoc tools (Nicholas & Nicholas, 1997).
Total RNA was extracted using the Trizol Reagent (Invitrogen) according to the manufacturer’s protocol. The total RNA concentration was quantified by spectro-photometer and 1 μg of total RNA was used for reverse transcribed into cDNA using First Strand cDNA synthesis kit (Roche). The amplification was performed with AmpliTag Gold DNA Polymerase (Applied Biosystems) in thermal Cycler (Bio-Rad) using the following parameters: denaturation at 95°C for 10 min and 30 cycles of reactions of denaturation at 98°C for 10 sec, annealing at 58°C for 30 sec, and elongation at 72°C for 45 sec. An aliquot of each PCR product was subjected to 1.2% (w/v) agarose gel electrophoresis and visualized by staining with ethidium bromide. The used PCR primers are listed in Table 1 and β-actin (ACTB) was used as an internal control.
RESULTS
Nam el al. isolated and identified all four TCR alpha, beta, gamma, and delta cDNAs and genomic clones from olive flounder leukocyte cDNA library (Nam et al., 2003). The sequences are 268 (PoTCR-alpha), 313 (PoTCR-beta), 279 (PoTCR-gamma), 322 (PoTCR-delta) amino acid residues in length. TCR subunits sequence alignment of the olive flounder are presented in Fig. 1. Amino acid sequence homology between TCR subunits is more than 80% and major structural difference is invisible between TCR subunits. Several important amino acid residues of TCR are conserved well in the variable region (W60, G67, D115, A117, Y119, C121).

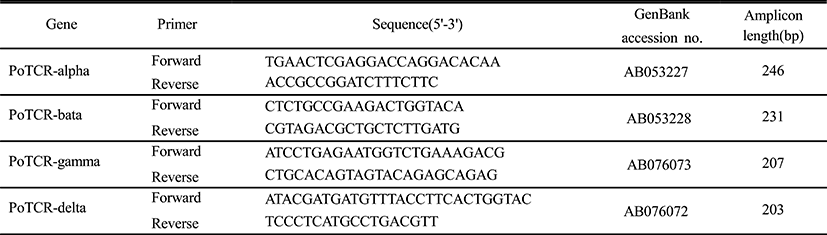
We examined the temporal expression of TCR subunits during the indicated early developmental time in the olive flounder. The expression of the TCR alpha subunit gene during the early stages of development is shown in Fig. 2. The mRNA expression of alpha subunit was already detected in the previous hatching step and expression level was maintained until metamorphosis at the same level. On the other hand, the expression pattern of another subunit is similar to the overall and expression of gamma and delta subunit were almost identical; the mRNA expressions were not observed during embryo development and the transcriptional level of TCR beta and gamma or delta increased for the first time at 1 dph and 0 dph, respectively. Also, the expression level was continuously maintained by a constant amount without increase or decrease until metamorphosis. Although the sequence homology is high, but there is functional difference between subunit. Therefore TCR subunits will important during the early stage to pathogen invasion in acquired immune response.
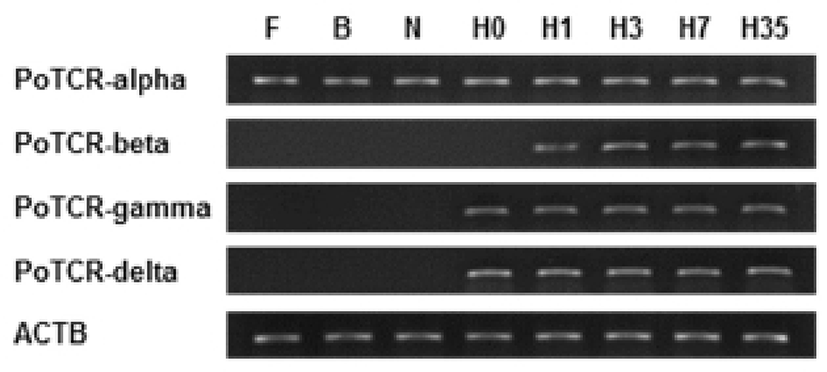
As a step toward evaluating function and localization of various olive flounder TCR molecules, we assessed the expression of the TCR subunit genes in various tissues from 2-year-old healthy olive flounder by RT-PCR. It was found that all TCR subunit mRNAs are significantly expressed in the spleen, kidney and gill. Also, transcript of TCR alpha and beta were expressed in the brain, fin, and eye and was not expressed in muscle. The overall pattern of mRNA expression of TCR gamma and delta was similar to alpha and beta, but was not expressed in the brain (Fig. 3).
Spleen, kidney and gill were typical immune organs in fish; kidney is a central organ with key regulatory functions for the immune-endocrine interactions. The spleen as a secondary lymphatic and scavenging organ plays a vital role in haematopoiesis, antigen degradation and antibody production processing (Rauta et al., 2012). Also, gill is the main mucosal surface and immune barriers. Therefore, expression of TCR subunits in immune organ can be expected that involved in the immune response.
DISCUSSIONS
The immune organs of fish homologues to the mammalian immune system. However, their structural complexity is less, potentially limiting the capability to generate fully functional adaptive immune responses against pathogen invasion (Tort et al., 2003). Moreover, the study of immune responses in fish is incomplete compared to human.
Lower vertebrates, like teleost fish, are the oldest animals with an adaptive immune system based on antibodies, B cells and T cells. (Flajnik & Kasahara, 2010). T cells play an essential role in cell-mediated immunity and as they interact with the bacteria in mucosal surfaces it seems they are very important in acquisition of tolerance or immunity against the infection (McGhee & Fujihashi, 2012). Although the value for the study of adaptive immune system is known to be high in fish, functional characterization of individual TCR subunits remains a challenging task.
In previous studies, TCR subunits sequence have been reported for various teleosts including rainbow trout, catfish, zebrafish, and Atlantic cod, olive flounder, bicolor damselfish, Atlantic salmon, carp, turbot, and puffer fish (Partula et al., 1996; Wilson et al., 1998; Hordvik et al., 2004); however, the information of each TCR subunit functional diversity in fishes has not been well documented.
In this study, to information of TCR subunits expression during early stage of development, we analyzed the expression pattern of TCR subunits at the three stages of embryos and five laval stages of days post hatching (Fig. 2). As a result, alpha subunit was expressed in the previous hatched stage, while the another subunits were increased at a later larval stage, although a few difference in time. The differential expression pattern between TCR subunits may indicate the functional difference in immunity. These results suggest that TCR subunits may have important roles in inhibiting the infection during early developmental stage.
The thymus is considered a key organ of the immune system in vertebrates. In this respect the thymus facilitates and regulates the interaction of lymphoid and non-lymphoid cells. This process is required because of lymphoid organs to guide and control the adaptive immune response through the very precise interaction between antigen presenting cells and lymphocytes. Early development of the thymus in fish has been studied in many diverse teleost species. The development of thymus affected temperature effects on growth and differentiation of the thymus occurs at the middle larval stage at 10 dph in olive flounder, but mRNA expression of TCR subunits was already detected from hatching stage. For such early expression, a role of TCR subunits before the differentiation of thymus in the development of the larvae independent of antigen presentation was suggested.
Also, we examined by RT-PCR using isolated various tissues from 2-year-old healthy olive flounder to investigate the tissue specific expression of the TCR subunit genes. The two different subunits making up the TCR heterodimer (alpha/beta and gamma/delta) were consistently expressed both in immune-related tissues (spleen, kidney, and gill), which are consistent with the function of T cell receptors. These results suggest that the olive flounder TCR subunits have differential transcriptional regulation in a broader range of tissues than found in mammals.
TCRs are heterodimers consisting of either alpha/beta or gamma/delta polypeptide combinations, and recognize MHC-presented epitopes. Therefore these molecules are useful markers for understanding the mechanisns of epitope recognition and tracing the migration of immune-related responses.
Although immune system of fish have been found highly conservation with mammalian in any part, it is difficult to clearly explain and proper application fish immunology based on the mammalian immune system. The egg and larval stages of marine fishes are the most critical in the successful survival and are extremely sensitive to environmental stress, especially pathogen infection (Rosenthal & Alderice, 1976). Therefore, further study is specifically required of the fish immune system to establish in its own the functions and characteristics, during early step of development which will provide a clear understanding of immune response.