INTRODUCTION
Lysozyme is one of the several antimicrobial enzymes, associated the first line immune defense parameter and is widely distributed in invertebrates and vertebrates. It is bactericidal in nature that catalyzes the hydrolysis of the β- 1,4-glycosidic linkage between N-acetylmuramic acid (NAM) and N-acetylglucosamine (NAG) alternating sugar residues in the bacterial peptidoglycan, and causes bacterial cell lysis (Bachali et al., 2002). The main sources of fish lysozymes are generally believed to be of leukocyte origin (monocytes, macrophages and neutrophils) (Lie et al., 1989; Cecchini et al., 2000).
In vertebrates, there are two types of lysozyme, chickentype (c-type) and goose-type (g-type) (Jollès & Jollès, 1984). In fish, c-type and g-type lysozymes have been identified from various species during last decades and lysozyme activity has been reported in different lymphoid tissues, kidney, gills and intestine (Saurabh & Sahoo, 2008). Ctype lysozymes have been isolated from many vertebrates and insects (Prager & Jollès, 1996, Qasba & Kumar, 1997). In teleost fish, lysozymes have been characterized in olive flounder (Hikima et al., 1997), brill (Jiménez-Cantizano et al., 2008), grass carp (Ye et al., 2010), orange-spotted grouper (Wei et al., 2012), and kelp grouper (Harikrishnan et al., 2011).
G-type lysozymes have also been characterized in four avian species, goose, chickens, black swans and ostriches (Simpson & Morgan, 1983; Nakano & Graf, 1991; Simpson et al. 1980; Schoentgen et al., 1982). The first report of a g-type lysozyme in fish was in olive flounder (Hikima et al., 2001), followed by other fish such as orange-spotted grouper (Yin et al., 2003), large yellow croaker (Zheng et al., 2007), Atlantic cod (Larsen et al., 2009), grass carp (Ye et al., 2010) and catfish (Wang et al., 2013). In a recent study, several studies was observed that up-regulation of g-type lysozyme was much stronger than ctype lysozyme after in vitro and in vivo stimulation with lipopolysaccharide (LPS) or challenge with bacterial pathogens, whereas c-type might be main molecule for housekeeping defense under normal conditions (Jimenez-Cantizano et al., 2008; Larsenet al., 2009; Ye et al., 2010).
Olive flounder (Paralichthys olivaceus) is recognized as economically important marine fish and successfully cultured in Japan, China and Korea. Recently, this species faces significant high mortality especially in early developmental stage, which attracted our interest to characterize how their immune system works during the development stages. In fishes, larvae are immediately exposed to various pathogens after hatching (Zapata et al., 1997). However, the immune system is still developing and not all of the structures and functions present in the adults are in the larvae (Ellis, 1988; Tatner, 1996). Therefore, knowledge of the immune system in ontogeny can offer new protective strategies and indicate optimal vaccination point (Vadstein, 1997).
Previous studies show that lysozyme has been detected in oocytes, fertilized eggs and larval stages of several fish species including tilapia (Brown et al., 1997), salmonids (Medzhitov et al., 2000), sea bass (Cecchini et al., 2000), Atlantic cod (Seppola et al., 2009) and rohu (Nayak et al., 2011). Although the lysozyme genes has been reported in several fish, considerably less is known about the expression of different types of lysozymes during early developmental stages in olive flounder. The expression study of lysozyme genes during ontogenesis will help in understanding the maturation and onset of humoral immunity during development of olive flounder immune system. Therefore, we investigate the expression pattern analysis of c-type and g-type lysozyme in early developmental stages, and tissue-specific expression study in immune organ.
MATERIALS AND METHODS
Larvae and adult flounder was collected from Genetics and Breeding Research Center, National Fisheries Research and Development Institute (NFRDI; Geoje, Republic of Korea), and maintained in 10 tons flow through tank at 20±1°C under a natural photoperiod. The tissue samples was prepared from various tissues including brain, muscle, fin, eye, liver, gill, kidney, and spleen obtained from healthy olive flounder (n=3). Eggs were collected from different developmental stages of embryo (~0.92 mm) including unfertilized oocyte (UFO), fertilized egg (F), 1- 16 cells (E1), molura (E2), blastula (E3), gastrula (E4), early neurula (E5), late neurula (E6), and lavae were collected from eight laval stages (~2.49 mm) including 0, 5, 10, 15, 20, 30, 40 and 50 days post hatching (dph) fish kept at 18.0°C in the sea-water tank. All the samples were collected in Trizol Reagent (Invitrogen) aseptically and preserved at –80°C until RNA extraction.
Total RNA was extracted from 50 to 100 mg using Trizol Reagent (Invitrogen), as per manufacturer’s instructions, and suspended in DEPC-treated water for RT-PCR. Total RNA concentration was quantified by spectrophotometer and 1-2 μg of total RNA was used for reverse transcribed into cDNA using First Strand cDNA synthesis kit (Roche). The amplification was performed with AmpliTag Gold DNA Polymerase (Applied Biosystems) in thermal Cycler (Applied Biosystems) using the following parameters: denaturation at 95°C for 10 min and 35 cycles of reactions of denaturation at 98°C for 10 sec, annealing at 58°C for 30 sec, and elongation at 72°C for 30 sec. An aliquot of each PCR product was subjected to 1.2% agarose gel electrophoresis and visualized by staining with ethidium bromide.
All the primers were designed using bundle software, Primer express (version 3.0). The gene specific primers are listed in Table 1.
To evaluate mRNA levels of c-type and g-type lysozyme, primers were specifically designed to detect and quantify cDNA sequences without detecting genomic DNA. The real-time PCR reactions were monitored with melting curve analysis using 7500 software (version 2.0.5). Amplification efficiency was determined by serial dilutions. All experiments were repeated in tri-replicate. Each reactions displayed an efficiency 92.9% (R2=0.982), 90.7% (R2=0.964) and 98.0% (R2=0.999), c-type, g-type and β-actin, respectively.
Real-time PCR reaction mixture contained 2 μL of diluted cDNA sample, specific primer and the FAST SYBR Green PCR Master Mix (Applied Biosystems). The reactions for the amplification of each genes was subjected to an initial denaturation at 95°C for 20 sec, annealing at 58°C for 30 sec, and elongation at 60°C for 30 sec. Thermal cycling and fluorescence detection were conducted using the 7500 Fast Real-Time PCR system (Applied Biosystems).
Expression of lysozyme genes was normalized to an endogenous reference β-actin and presented as subtraction of target CT values from β-actin CT values (ΔCT value). Comparison of gene expression between tissues and calibrator was derived from subtraction of the calibrator ΔCT values from the target ΔCT values to give a ΔΔCT value, and relative gene expression was calculated to determine fold difference (2-ΔΔCT).
The data are presented as the mean ± standard deviation. The mRNA levels of lysozyme genes were expressed as a ratio to those β-actin, which was simultaneously amplified as an internal control for each cDNA. The data was statistically analyzed by one-way ANOVA after arcsine transformation when needed, and followed by a Tukey’s test for identification of the statistically distinct groups. Significant differences were accepted for P <0.05.
RESULTS
In previous study, c-type lysozyme gene was expressed mainly in kidney, spleen, brain and ovary (Hikima et al., 1997), whereas the g-type was detected in kidney, spleen, skin, muscle, heart and brain (Hikima et al., 2001) using northern blot analysis. Such results are now confirmed by the detection of lysozyme in various tissues by real-time PCR analysis of olive flounder.
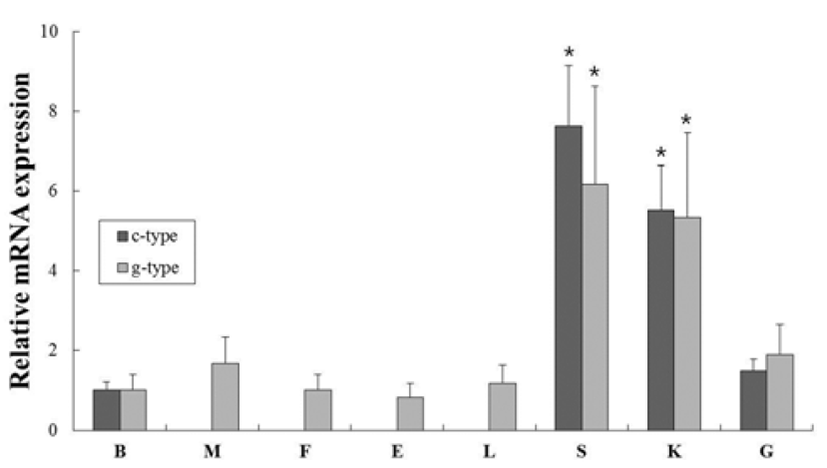
In order to investigate of c-type and g-type lysozyme mRNA expression in various tissues from healthy flounder, oligonucleotide specific primers were designed according to previously reported both genes in olive flounder (GenBank accession no. AB050469 and AB050591, respectively) (Table 1). Gene expression levels were normalized with respect to β-actin and brain mRNA was employed as calibrator. C-type lysozyme was highly expressed in spleen, kidney, gill and brain (7.6-, 5.5- and 1.5-fold higher than in the brain, respectively). In contrast, g-type lysozyme was expressd ubiquitously in transcript level in all tissues. The highest level was expressed in spleen and kidney (6.2- and 5.3-fold higher than in the brain, respectively). These overall patterns of lysozyme genes were highly expressed in spleen and kidney, similar to previous studies (Fig. 1). The spleen and kidney (anterior and middle) are one of largest lymphoid organs and phagocytic organ in teleost fish (Zapata et al., 2006).
All samples were technically triplicated samples, and target genes were standardized using reference gene β- actin for mRNA expression study. The expression pattern of the c-type and g-type lysozyme genes were investigated over time course UFO, F, E1 to E6 (egg stages) and 0 to 50 days post hatching (larval stages) of olive flounder using real-time PCR.
In our study, the c-type lysozyme mRNA transcripts were detected in unfertilized oocytes (UFO), but not detected in g-type lysozyme (Fig. 2). Studies on several fish species have shown that lysozyme genes are transferred from the mother to the offspring (Yousif et al., 1994, 1994; Wang & Zhang, 2010), and it may be involved in the early defense against pathogen in eggs containing developing embryo and larvae.
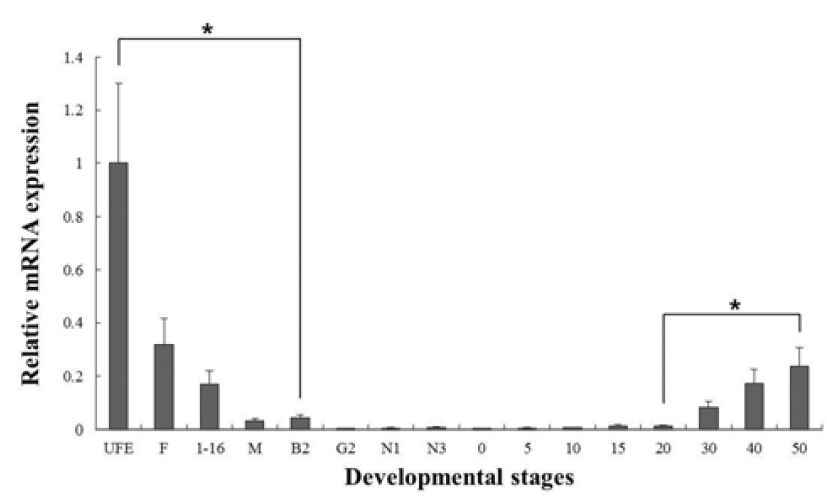
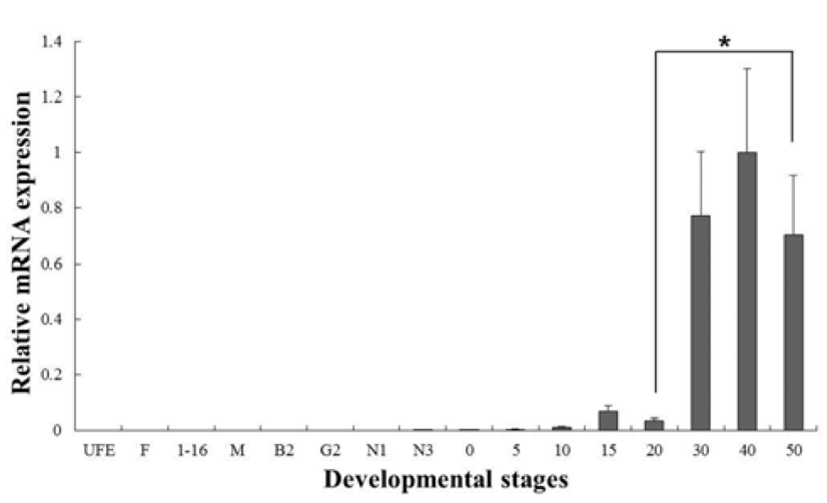
C-type lysozyme expression was highly detected at unfertilized oocyte (UFO), and significantly decrease until around hatching stage (0 dph). After hatching, transcription level was significantly increased in 30 to 50 dph (Fig. 3). G-type lysozyme mRNA transcripts were first detected at late neurula (E6), and the transcription level was drastically increased after the 20 dph. The intensity of g-type lysozyme transcript was measured on the basis of E6 (1.0- fold), each value has the 30 dph (1583-fold), 40 dph (2051-fold) and 50 dph (1445-fold), respectively (Fig. 4).
DISCUSSIONS
The fish immune system is not fully developed during early embryonic stages and the immunological capacity is thus assumed to be limited. C-type and g-type lysozyme genes have revealed as useful markers to study the effect of environment contaminants (Mondon et al., 2000; Skouras et al., 2003; Nakayama et al., 2007), immunostimulants (Ashihida & Okimasu, 2005; Alvarez-Pellitero et al., 2006) as well as bacterial pathogens (Hikima et al., 2001; Minagawa et al., 2001) on the immune response in teleost fish. Therefore their levels of expression can help from the problem of high mortality in olive flounder larvae.
In our study, c-type and g-type lysozyme was investigated in various tissues including brain, muscle, fin, eye, liver, gill, kidney, and spleen (Fig. 1). The results are expressed predominantly in hematopoietic tissues (spleen and kidney). In previous study, fish lysozyme is generally believed to be of leucocyte origin (Lie et al., 1989; Cecchini et al., 2000), and localization of lysozyme in prelarvae, before the onset of feeding, has been studies in tilapia using immunohistochemistry (Takemura, 1996). In many species, lysozymes seem to be ubiquitously expressed in most tissues. However, few studies of lysozymes are reported that is specifically expressed. In cows, c-type lysozymes are differentiated by their function and tissue expression patterns as stomach and non-stomach lysozymes (Irwin 1996, 2004). Moreover, in zebrafish (Liu & Wen, 2002) and in rohu (Nayak et al., 2011), those are specifically expressed in the myeloid lineage and hematopoietic tissues, respectively. The g-type lysozyme is also reported with tissue-specific expression in chicken, whose g-type lysozyme gene is expressed in bone marrow and lung (Nakano et al., 1991).
In several studies, the patterns of lysozymes expression level, they did not follow the trend of an initial increase in larval stage. Therefore, we have included the investigations of maternal mRNA transfer into oocytes in unfertilized eggs. As a result, we found that expressing of c-type lysozyme mRNA transcripts in unfertilized oocytes, but not expressed in g-type lysozyme (Fig. 2). Also, c-type lysozyme mRNA was first detected in unfertilized oocyte (UFO) stage, observed the significantly decreased mostly until hatching stage (H), and was significantly increased in 30 to 50 dph after hatching stage (Fig. 3). On the other hand, gtype lysozyme mRNA transcripts were first detected at late neurula (E6) stage, and the mRNA level was significantly increased after 20 dph (Fig. 4). In zebrafish, maternally supplied mRNAs are selectively degraded prior to the activation of embryonic transcription. This occurs in order to transfer developmental control to the zygote genome and a correctly timed clearance of maternal transcripts is critical for embryogenesis (Giraldez et al., 2006). After hatching stages, the mRNA level of c-type lysozyme was increased significantly indicating that from this period the embryo independently produces its own transcripts and the use of maternally derived lysozyme may be of less importance.
In summary, this study provides new information of lysozymes mRNA expression during the early developmental stage in olive flounder. Although it is still not clear whether presence of components of the immune system confer any protection against pathogens, maternal supply and early expression may indicate an important function during the early stages of development in olive flounder.
