INTRODUCTION
The C-terminal domain (CTD) of RNA polymerase II (RNAPII) plays a crucial role in the transcriptional regulation and RNA processing of protein-coding genes. Additionally, substantial evidence suggests that the CTD is involved in the transcription and processing of spliceosomal U small nuclear RNAs (U1 to U5 snRNAs) dependent on RNAPII. The integrator complex (INT) is responsible for processing the 3′-end of spliceosomal snRNAs and controls the transcription of protein-coding genes by facilitating transcription termination following unstable pausing. In 2005, Baillat et al. made this discovery as part of their initiative to identify interacting partners of DSS1 (Deleted in split hand/split foot1), a protein linked to congenital limb defects in humans (Baillat et al., 2005). INT is absent in lower eukaryotes, such as yeast, but is conserved in higher eukaryotes (Sabath et al., 2020). It comprises at least 14 subunits in mammalian cells, and for the majority of these subunits, both their function and structure remain uncharacterized. (Welsh & Gardini, 2023).
After the discovery of the integrator, it has become increasingly apparent that this complex can regulate the processing and expression of RNA PolI transcripts beyond snRNAs. These transcripts include a range of nonpolyadenylated RNAs, such as enhancer RNA (eRNA), telomerase RNA, viral miRNAs, and replication-dependent histone, along with the transcription of numerous canonical protein-coding genes and long noncoding (lnc) RNAs (Baillat et al., 2005). The dysregulation of integrator expression has been associated with developmental and disease phenotypes in various metazoan species. This correlation is not unexpected, as mutation in the integrator can result in the dysregulation of the expression of hundreds to thousands of mRNAs. Regarding the role of IntS13 in human myeloid differentiation, an increasing body of research indicates that integrator subunits play an essential role in normal development across metazoans (Mendoza-Figueroa et al., 2020). The mutation in the integrator complex subunit 1 gene (c) and subunit 8 gene (INTS8) in humans have been demonstrated to induce a severe neurodevelopmental syndrome. This syndrome is characterized by profound intellectual disability, epilepsy, rigidity, facial and limb dysmorphisms as well as subtle structural abnormalities in the brain (Peart et al., 2013; Oegema et al., 2017; Krall et al., 2019). In addition, the deletion or mutation of integrator subunits in Mus musculus (Ints1), Oryzias latipes (Ints1), Drosophila melanogaster (IntS4 and 7), Caenorhabditis elegans (Ints6), and Danio rerio (IntS6) have all been reported to result in abnormalities in early development and subsequent lethality (Chen et al., 2012; Rienzo & Casamassimi, 2016; Mendoza-Figueroa et al., 2020).
In vivo/in vitro model system with the targeted gene deleted provide researchers an important tool for studying gene function. Genome editing technologies, such as ZFN, TALEN and CRISPR/Cas9 systems, have emerged as powerful tools for characterizing gene functions, holding significant potential for applications in in human gene therapy (Ansai & Kinoshita, 2014). Depletion of integrator subunits, including IntS12, has a minimal impact on the 3′-end processing of snRNAs in both Drosophila and human HeLa cells (Mendoza-Figueroa et al., 2020). Therefore, creating an INTS14 knockout animal model is highly significant for studying its impact on the INT in transcription and developmental processes.
In this study, we initially examined the expression patterns of the ints14 gene in various cell types and tissues during developmental stages using zebrafish. We generated for the first time an ints14 mutant in zebrafish, using CRISPR/Cas9 technology, as a model to study developmental disease. A zebrafish mutant model for ints14/vwa9 gene would be an effective tool to investigate the underlying mechanisms during the processing and expression of RNA PolII with integrator-related genes in the developmental phenomenon.
MATERIALS AND METHODS
Zebrafish were maintained as described in a previous study (Aoyama et al., 2015). Wild-type embryos were obtained by breeding males and females. All animal studies were approved by the Animal Welfare and Research Ethics Committee at the National Marine Biodiversity Institute of Korea (MAB-23-03), and all procedures were conducted strictly in accordance with the Guide for the Care and Use of Laboratory Animals. All surgery was performed under anesthesia, and every effort was made to minimize animal suffering.
Total RNA was extracted using the TRIzol reagent (Ambion, Austin, TX, USA), and the RNA was quantified with a spectrophotometer (Qiagen, Hilden, Germany). First-strand cDNA was synthesized from total RNA (1 μg) with a SuperScript IV first-strand Synthesis Kit (Invitrogen, Carlsbad, CA, USA). The ints14 cDNA was amplified with real-time PCR, using the ints14 forward primer (5′-CATAACTGTTCTTCAGCGTGTAAA-3′) and ints14 reverse primer (5′-CACTCCTGCTGTACTACATTACT-3′). Beta-actin cDNA was amplified with PCR, using the b-actin forward primer (5′-CGAGCTGTCTTCCCATCCA-3′) and b-actin reverse primer (5′-TCACCAACGTAGCTGTCTTTCTG-3′). The reaction conditions were as follows: 95℃, 5 min; 33 cycles of 95℃ for 30 s, 56℃ for 30 s, and 72℃ for 1 min; and then 72℃, 5 min. PCR produce size was estimated via electrophoresis on a 1.2% agarose gel.
Whole-mount in situ hybridization (WISH) was performed as previously described (Jung et al., 2012). Antisense DIG-labeled riboprobes for INTS14 were synthesized from linearized plasmids by using Sp6 RNA polymerase (Ambion). Photos images were taken with a stereo zoom microscope (SMZ25, Nikon, Tokyo, Japan).
sgRNA (5′-ACGCGACCGGTGTCAGTGGAGGG-3′) of ints14 (accession number. NM_199834) was generated and designed by using Toolgen (Seoul, Korea). Cas9 protein was purchased from Toolgen. Ints14 target sequences are located in exon 2. Ints14 sgRNAs (500 ng per 1 μL) and Cas9 protein (1 μg per μL) with 2 μL of 0.5% phenol red were mixed and injected into 1-cell stage embryos, which were subsequently raised to adulthood.
Adult zebrafish were anesthetized with 0.6 mM Tricaine solution (Sigma-Aldrich, St. Louis, MO, USA). Zebrafish caudal fins were amputated with a surgical scalpel blade (Cat. HSB 704-10, Hammacher, Solingen, Germany). For genomic DNA preparation, amputated caudal fins of adult fish or whole single embryos at 1–3 dpf (day post-fertilization) were lysed essentially as described [18]. The lysates [50 mM NaOH, 1M Tris-HCl (pH 8.0)] were used for PCR amplification.
PCR amplification of the targeting sequences in the ints14 gene for 436 bp PCR products was performed with HiPi DNA polymerase (Elpis, Daejeon, Korea) via PCR using the following primers: ints14 exon 2 forward primer (5′-CATAACTGTTCTTCAGCGTGTAAA-3′) and ints14 exon 3 reverse primer (5′-CACTCCTGCTGTACTACATTACT-3′). The reaction program comprised the following steps: 95℃ for 5 min, 33 cycles of 95℃ for 30 s, 56℃ for 30 s, and 72℃ for 1 min, followed by 72℃ for 5 min. The PCR products were gel purified and assayed using T7E1 enzyme (Toolgen), cloned into the pGEM T-easy vector (Promega, USA), and sequenced to determine the exact sequences (Macrogen, Seoul, Korea).
RESULTS AND DISCUSSION
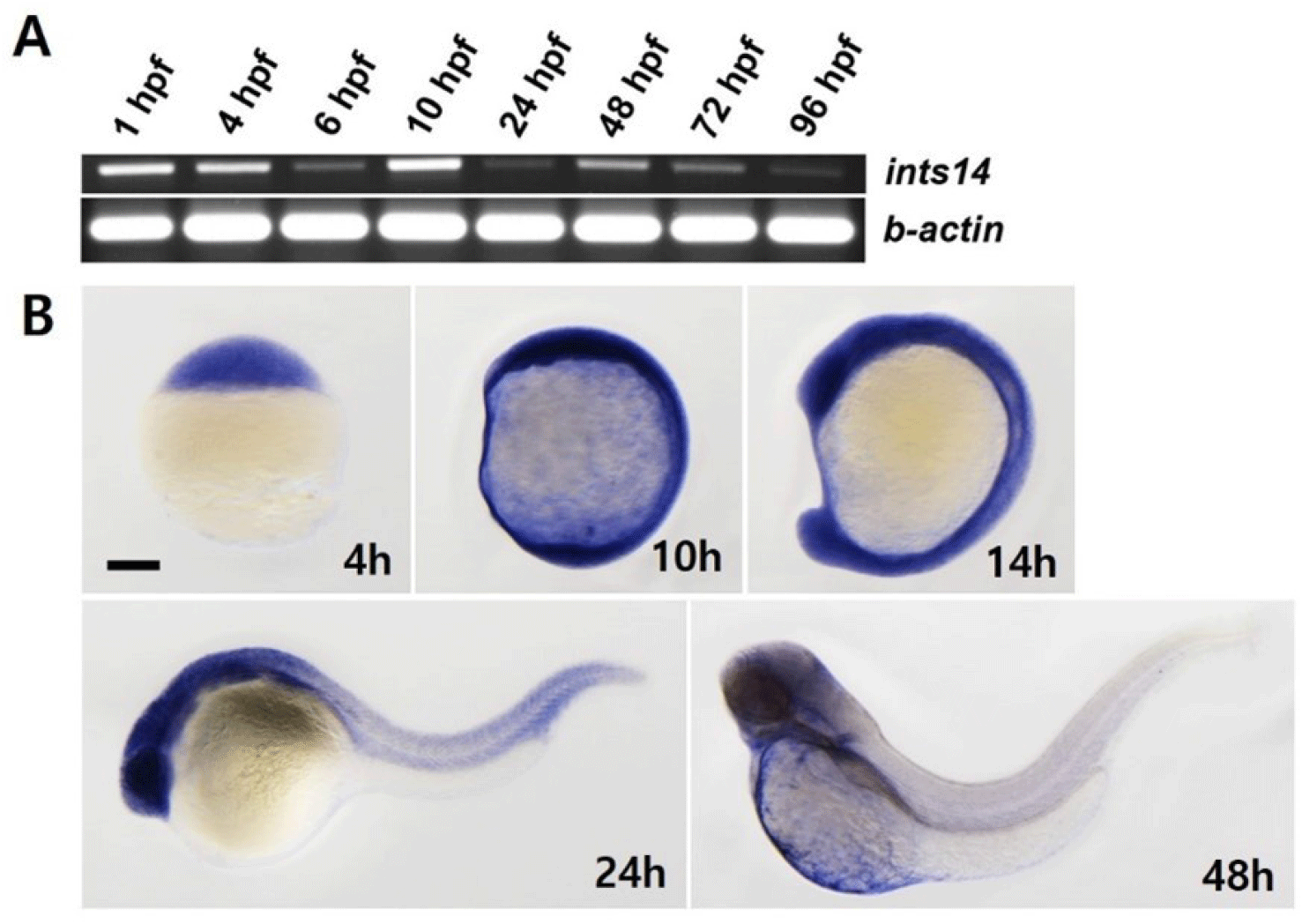
We aimed to observe the spatiotemporal expression of ints14 at different stages of zebrafish embryo development by using reverse transcription polymerase chain reaction (RT-PCR) and WISH. As shown in Fig. 1A, ints14 expression, detectable in the maternal gene expression stage, remained constant from 6 to 96 hpf (hour post fertilization). WISH data showed that ints14 mRNA was expressed ubiquitously from the blastula stage to segmentation (Fig. 1B). In 24 hpf embryos, ints14 was strongly expressed in the brain and eye and weakly expressed in the trunk and tail region. In 48 hpf embryos, ints14 was expressed in the brain and yolk sac. At 48 hpf, ints14 expression was maintained in the brain. Spatiotemporal expression pattern analysis showed that ints14 functions in various cells from the early developmental stage. Thus, it can be confirmed that ints14 affects brain development at later stages.
The INTS14/VWA9 protein is a multifunctional protein complex that consists of 14 subunits referred to as Int1–Int14 (INT subunits). This complex plays a crucial role in determining the fate of numerous nascent RNAs transcribed by RNAPII. Multiple individuals with severe neurodevelopmental delay and cancers that carry biallelic mutations in integrator subunits have now been identified (Mendoza-Figueroa et al., 2020). Genetic disease models in the ints14 gene have not been reported to date. Thus, there is an unmet need to generate ints14 gene knockout zebrafish to model this phenomenon. The zebrafish ints14 gene (GenBank accession number: NM_199834.1) is located on zebrafish chromosome 18. Twelve exons have been identified, with the ATG start codon in exon 2 and the TAG stop codon in exon 12. Exon 2 was selected as a target site. We designed a sgRNA targeted on exon 2 of the zebrafish ints14 genome (Fig. 2, Table 1). It appears that the designed and produced target sequence sgRNA containing the NGG protospacer adjacent motif required for Cas9 cleavage will not have the activity of binding to any other site in the zebrafish genome.
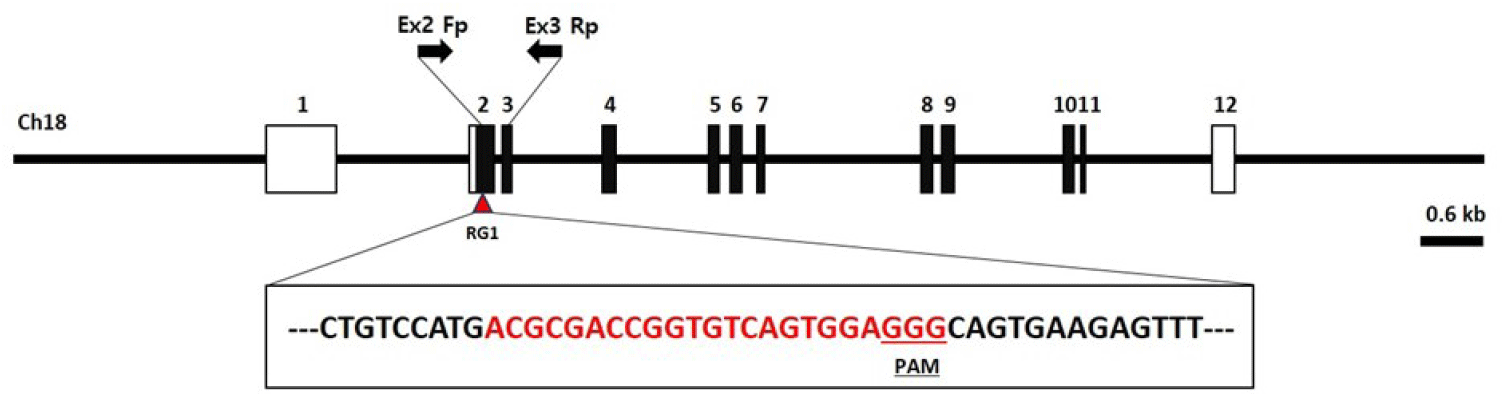
| Gene | Accession number | RGEN name | Sequence (5′-3′) | Region |
|---|---|---|---|---|
| ints14 | NM_199834 | RG1 | ACGCGACCGGTGTCAGTGGAGGG | Exon 2 |
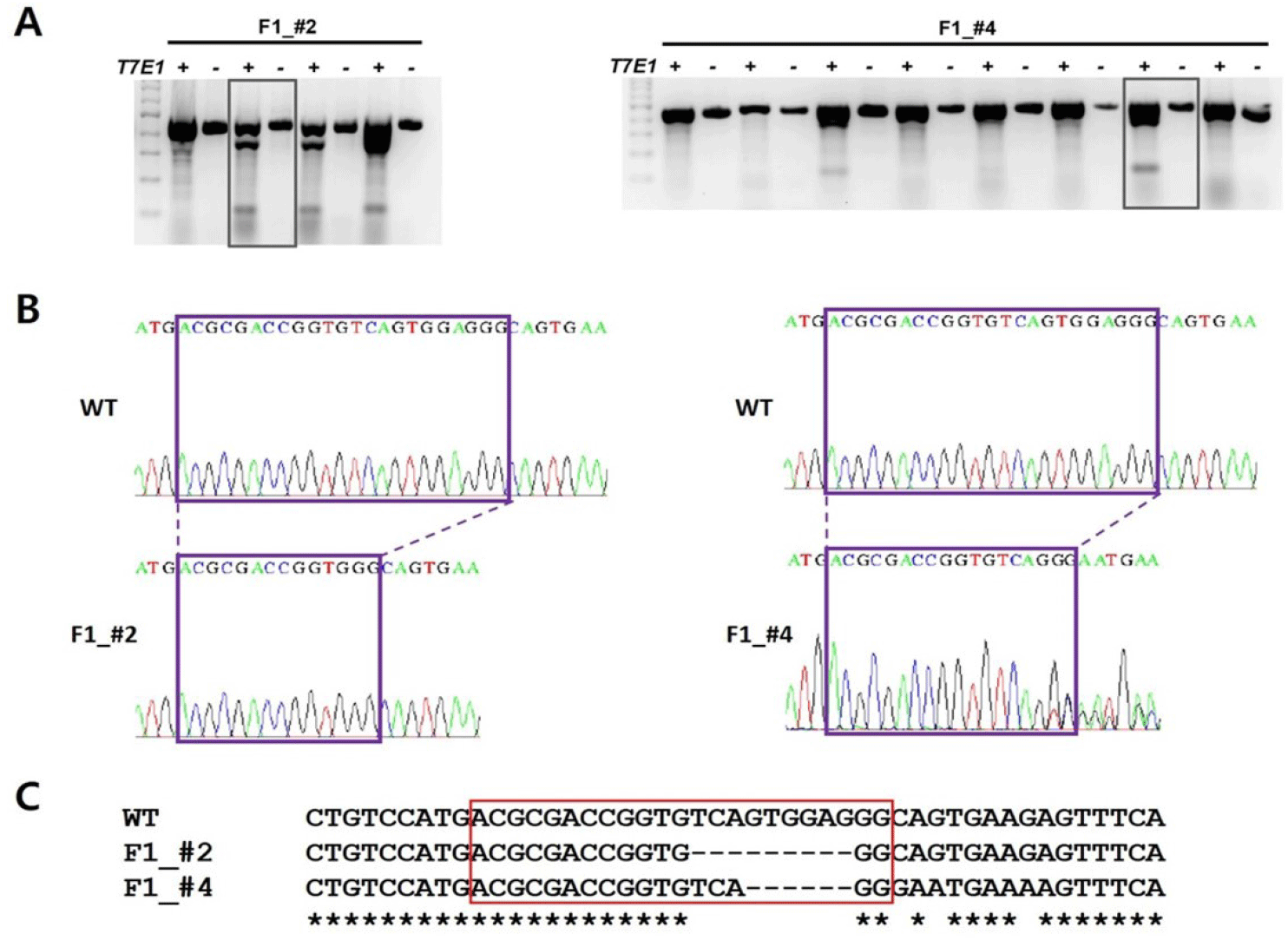
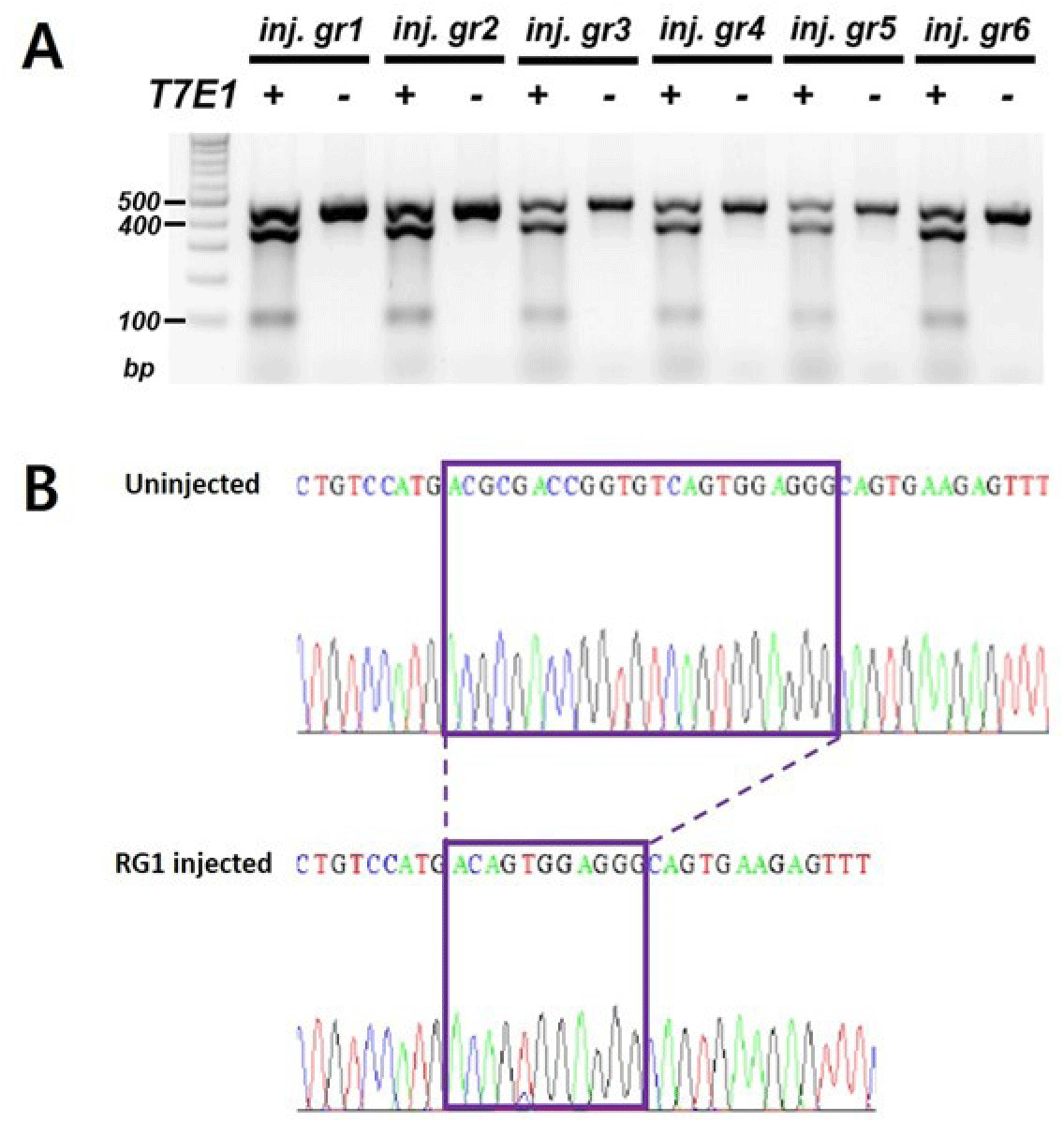
To generate ints14 zebrafish mutant utilizing CRISPR/Cas9 technology, we tested the efficiency for sgRNA by co-injecting with Cas9 protein into one-cell stage embryos. After performing DNA sequencing and T7E1 assay for mismatch of target sequences in co-injected embryos, we found that the sgRNA efficiently generated mismatched DNA for the target region, suggesting successful generation of the ints14 mutant zebrafish (Fig. 3, Table 2).
| Target of sgRNA | PCR size | T7E1 assay | ID primer | Sequence | |
|---|---|---|---|---|---|
|
ints14 exon2 |
436 bp | 336±@ bp | 100±@ bp | Ex2 Fp | CATAACTGTTCTTCAGCGTGTAAA |
| Ex3 Rp | CACTCCTGCTGTACTACATTACT | ||||
The Cas9 protein and single sgRNA were co-injected into the cytoplasm of zebrafish fertilized eggs. Founder fishes injected with ints14 and Cas9 were crossed with wild-type to obtain 1–3 days embryo. To confirm the disruption of ints14 gene in these fish, PCR and T7E1 assays were performed (Table 2). Compared with wild-type zebrafish, the desired fragment deletion of ints14 gene spanning the sgRNA site was detected by using T7E1 assay. A clear band demonstrates the fragment deletion of ints14 in #2 and #4 lines of the zebrafish F1 generation (Fig. 4A). The TA-cloning and sequencing results demonstrate that a deletion of approximately 9 or 6 bp can be achieved via the single gRNA system (Fig. 4B and C). These frameshift mutations of ints14 resulted in termination site change and early termination of the INTS14 protein, indicating that the INTS14 protein was abnormally generated. However, no lethality phenomenon was observed in the early embryonic stages of the ints14 knockout model. In the future, there is a need for research on behavioral and developmental abnormalities during the later stages. While the molecular mechanisms of ints14 have been reported, future research is anticipated to explore its cellular and developmental functions using the model we have established. The investigation into the interplay of INTS14 with cellular processes and its potential associations with human diseases holds promise for advancing our understanding of its broader implications.