INTRODUCTION
Mouse pluripotent stem cells are generally classified into two interchangeable states: naïve pluripotency and primed pluripotency, are represented by embryonic stem cells (ESCs) and epiblast stem cells (EpiSCs), respectively (Nichols & Smith, 2009). ESCs are derived from the inner cell mass (ICM) of blastocyst-stage embryos and are characterized by a dome-shape, dense cluster morphology (Boroviak et al., 2014; Martello & Smith, 2014). In contrast, EpiSCs are derived from epiblast at E5.5-7.5 after implantation and exhibit a flat, monolayer-spreading cluster morphology (Najm et al., 2011; Kojima et al., 2014). Although both refer to pluripotency, they significantly differ in various biological characteristics, including morphology, gene expression patterns, maintenance conditions, epigenetic status, chimerism contribution ability, and germ cell differentiation ability (Tsukiyama & Ohinata, 2014; Yu et al., 2021). In particular, EpiSCs have been reported to resistant to differentiation into primordial germ cell-like cells (PGCLCs) (Hayashi et al., 2011), suggesting that EpiSCs are already in a developmentally biased state. In comparison, epiblast-like cells (EpiLCs) mimic the state of the upper germ layer at E5.75 in vivo and are highly responsive to PGC-inducing cytokines including bone morphogenetic protein 4 (BMP4), allowing them to effectively differentiate into PGCLCs (Hayashi et al., 2011; Nakaki et al., 2013). Although both EpiSCs and EpiLCs belong to a primed pluripotent state, EpiLCs are known to have a higher reprogramming ability and responsiveness to PGCLC induction because they mimic a narrower developmental time point reflecting the epithelial epiblast before gastrulation (Endoh & Niwa, 2022).
PGCs, the embryonic precursors of germ cells (PGC), are a key cell population for understanding reproductive development and germ cell transmission. In the mouse, at about E6.0, signals from the adjacent extraembryonic ectoderm initiate the induction of potential precursors of the PGCs within the proximal epiblast cell population. The formation of final PGCs is induced in the epiblast at about E7.25. At that time about 30 cells acquire the PGC fate (Ginsburg et al., 1990; Lawson et al., 1999; Saitou et al., 2002). In vitro derivation of PGCLCs provides a powerful platform to simulate early germ cell development, suggesting promising applications in reproductive biology and regenerative medicine. Traditionally, PGCLCs have been induced from intermediate EpiLCs stage under defined cytokine conditions, particularly BMP4 signaling (Hayashi et al., 2011). Nevertheless, EpiSCs can be stably cultured for long periods of time, and they have the advantage of being an in vitro model that reflects the late epiblast state in vivo (Endoh & Niwa, 2022). Therefore, studies that improve the conditions for inducing EpiSC-derived PGCLCs will provide an experimental platform for evaluating the germ cell fate determination ability in a more developmentally diverse pluripotent state.
Recently, Yu et al. have shown that PGCLC specification can also be achieved directly in EpiSCs, but the efficiency of PGCLC induction from EpiSCs remains relatively low, and the optimal conditions for enhancing this process have not yet been fully established (Yu et al., 2021). Most existing studies have focused on PGCLC induction from EpiLCs, leaving a significant lack of systematic comparative analysis of efficient EpiSC-based induction strategies. In particular, there have been few attempts to directly compare the effects of culture environment (2D vs. 3D) in EpiSC-based PGCLC induction or to improve induction efficiency by applying 3D conditions. In this study, we hypothesized that PGCLC induction from mouse EpiSCs would be more efficient in a 3D aggregate environment than in conventional 2D monolayers. To test this, we used a transgenic Blimp1-mVenus×Stella-ECFP (BVSC) reporter ESC to track PGCLC formation and compared differentiation outcomes between 2D and 3D culture conditions. By quantifying double-positive PGCLCs by flow cytometry, we aimed to elucidate the role of spatial organization in promoting germ cell fate. Our results demonstrate that 3D aggregates significantly enhance PGCLC yield from EpiSCs, emphasizing the importance of culture dimension in recapitulating in vivo-like developmental cues. This study contributes to improving PGCLC differentiation protocols and provides insight into environmental factors that influence germ cell differentiation from primed pluripotent stem cells.
MATERIALS AND METHODS
The following reagents and chemicals were purchased from respective suppliers: Dulbecco’s modified Eagle’s medium (1×) [DMEM; Gibco (Grand Island, NY, USA), 11965-092], DMEM/F12 (Gibco, 11320-033), Neurobasal (Gibco, 21103-049), fetal bovine serum (FBS; Gibco, 16000-044), Pen-Strep-Glutamine (100×) (PSG; Gibco, 10378-016), N2 supplement (Gibco, 17502-048), B27 supplement (Gibco, 12587-010), 2-Mercaptoethanol (55 mM; Gibco, 21985-023), Glutamax (100×) [Thermofisher (Waltham, MA, USA), 32571-036], Non-essential amino acid (100×) (Thermofisher, 11140-050), Na pyruvate (Gibco, 11360-070), LIF (Sigma-Aldrich, ESG1107), bFGF (Thermofisher, 13256-029), IWR1 [Sigma-Aldrich (St. Louis, MO, USA), I0161], BMP4 [Peprotech (Cranbury, NJ, USA), 120-05ET], PD0325901 [Stemgent (Cambridge, MA, USA), 04-0006], CHIR99021 (Stemgent, 04-0004), Phosphate-buffered saline (PBS; Gibco, 10010-023), Trypsin-EDTA, 0.25% (Gibco, 25200-072), Paraformaldehyde [Biosesang (Seongnam-si, Gyeonggi-do, Korea), PC2031-050-00], Triton X-100 (Sigma-Aldrich, 282103), BSA (Sigma-Aldrich, A8806), Accutase [Biolegend (San Diego, CA, USA), 42301], Y27632 (Peprotech, 1293823).
The Blimp1-mVenus Stella-ECFP (BVSC) transgenic female (XX) H18 ES cell line (Acc. No. BV, CDB0460T; SC CDB0465T: http://www.cdb.riken.jp/arg/TG%20mutant%20mice%20list.html) was received from Saitou (Hayashi et al., 2011, 2012). ESCs were cultured in N2B27 medium supplemented with 3 μM CHIR99021, 1 μM PD0325901 and 1,000 U/mL LIF on gelatin-coated plates (N2B27+2iL). The N2B27 medium comprised 50% DMEM/F12, 50% Neurobasal, 1×N2, 0.5×B27, 1% PSG and 0.1mM 2-mercaptoethanol. The ESCs were passaged every 2 days by dissociating with 0.25% Trypsin-EDTA and cultured in a 37℃ 5% CO2 humidified incubator. All experiments were carried out using cells cultured within 10–30 passages (Ying et al., 2008; Kim et al., 2022).
EpiSC were generated by continual culture in N2B27 medium supplemented with 20 ng/mL bFGF and 2.5 μM IWR1 (FR medium) on MEF plates. EpiSCs were passaged using Accutase and seeded as single cells at approximately 5×104 cells in a well of a 6-well plate every 3d. The medium was changed daily (Yu et al., 2021).
EpiSCs were dissociated into single cells using Accutase and plated at a density of 5×103 cells per well of 24-well plate coated with FBS in FR medium supplemented with 5 μM Y27632. The next day, FR medium was replaced with the iCD1 medium (DMEM medium supplemented with 0.5×N2, 1×B27, 1% GlutaMAX, 1% NEAA, 1% sodium pyruvate, 0.1 mM 2-mercaptoethanol, 50 μg/mL vitamin C, 10 ng/mL bFGF, 5 mM LiCl, 1,000 U/mL LIF, 1 μM CHIR99021, 10 ng/mL BMP4, 1 μM EPZ5676, and 2.5 μM EPZ6438). Medium was refreshed daily. After 3 days, iCD1 medium was changed into N2B27+2iL (described above). The medium was changed every day (Yu et al., 2020).
EpiSCs were dissociated into single cells using Accutase and plated at a density of 3×103 cells per well of u-bottom 96-well plate in FR medium supplemented with 5 μM Y27632. The next day, FR medium was replaced with the iCD1 medium (DMEM medium supplemented with 0.5×N2, 1×B27, 1% GlutaMAX, 1% NEAA, 1% sodium pyruvate, 0.1 mM 2-mercaptoethanol, 50 μg/mL vitamin C, 10 ng/mL bFGF, 5 mM LiCl, 1,000 U/mL LIF, 1 μM CHIR99021, 10 ng/mL BMP4, 1 μM EPZ5676, and 2.5 μM EPZ6438). Medium was refreshed daily. After 3 days, iCD1 medium was changed into N2B27+2iL (described above). The medium was changed every day (Yu et al., 2020).
For the FACS analysis, the PGCLCs cultured on 2D or 3D culture plates were harvested and transferred to a 19 mL conical tube. Then cells were dissociated into single cells with 0.25% Trypsin-EDTA and neutralized with DMEM containing 15% FBS. After the dissociation, cells were collected by centrifugation (205×g for 5 min at 4℃) and resuspended with flow cytometry buffer (PBS supplemented with 1% FBS). The samples were analyzed using a High-end Performance Flow Cytometry system (FACSymphony A3, Becton Dickson, Fraklin Lakes, NJ, USA) installed at the Center for University Research Facility (CURF) at Jeonbuk National University (Kim et al., 2022). The gating strategy was as follows: (1) debris was excluded based on FSC-A and SSC-A profiles; (2) single cells were selected by FSC-A versus FSC-H gating; (3) PGCLC populations were identified based on Venus (Blimp1) and CFP (Stella) fluorescence intensity using the BVSC reporter system.
The cells were seeded fixed with 4% paraformaldehyde for 30 min at room temperature. The cells were washed with PBS once and subsequently soaked in the mixing buffer of equal volume of 0.1% Triton X-100 and 3% BSA for 1 h at room temperature. The cells were washed with PBS once, then incubated with primary antibody at 4℃ for overnight. The cells were washed with PBS five times next day, then incubated with appropriate secondary antibody for 1 h at room temperature. The cells were washed with PBS five times to remove the secondary antibody and counterstained with 4’,6-diamidino-2-phenylindole (DAPI) for 3 min and washed with PBS once. Primary antibodies and secondary antibodies were diluted in 3% BSA. Primary antibody used in this study was anti-Nanog (Bethyl Laboratories (Montgomery, TX, USA), A300-397A, 1:500) (Jang et al., 2024). To quantify BVSC-positive cells, we performed DAPI nuclear counterstaining and calculated the proportion of BVSC-positive cells over total DAPI+nuclei using ImageJ version 1.53t (National Institutes of Health, Bethesda, MD, USA).
To assess the relative gene expression level, RNA was extracted from cells using an AccuPrep® Universal RNA Extraction Kit [Bioneer (Daejeon, Korea), K-3141] following the manufacturer’s protocol. Total RNA (1 μg) was reverse transcribed with an Accupower® CycleScript RT Premix (Bioneer, K-2047-B) according to the manufacturer’s instructions. Relative gene expression was measured in triplicate using Powerup SYBR Green Master Mix (Applied Biosystems, A25918). For the examination of expression level, the gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was employed as an internal control. Reactions were run on the LightCycler® 96 system, and results were analyzed as 2−ΔΔCt normalized to gene expression (Kim et al., 2022). The primers used for qRT-PCR were as follows: GapDH sense 5’-ATGAATACGGCTACAGCAACAGG-3’, GapDH antisense 5’-CTCTTGCTCAGTGTCCTTGCTG-3’, Stella sense 5’-AGGCTCGAAGGAAATGAGT TTG-3’, Stella antisense 3’-TCCTAATTCTTCCCGATTTTCG-5’.
All experiments were performed in triplicate, and the data of all repetitions of each experiment were collated and expressed as means±SE of the mean. Statistical tests were conducted using SAS version 9.4 (SAS Institute, Cary, NC, USA), and statistical differences were analyzed using the Student’s t-test or analysis of variance (ANOVA), followed by Duncan’s Multiple Range Test for post hoc comparisons. A p-value<0.05 was regarded as significant.
RESULTS
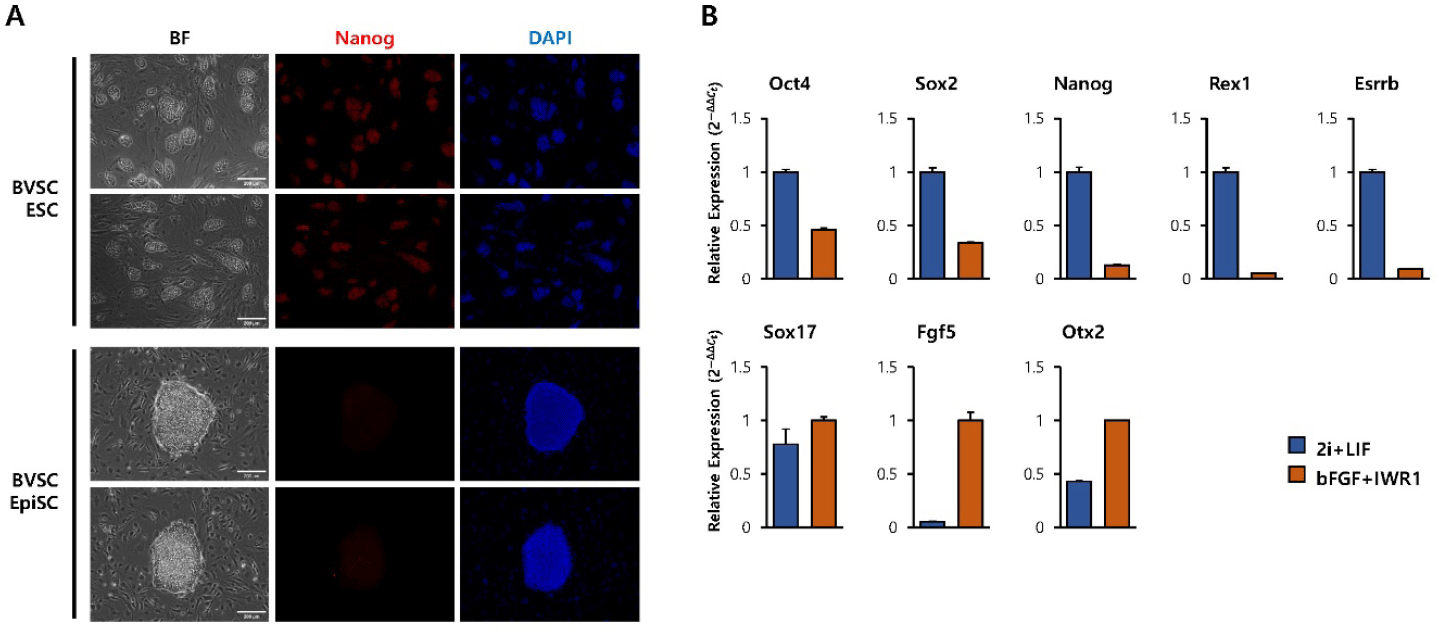
To generate primed pluripotent EpiSCs, BVSC transgenic ESCs were cultured under EpiSC maintaining conditions on MEF feeders (Yu et al., 2021). The differentiated cells exhibited a flat, epithelial-like monolayer morphology, which was distinct from the dome-shaped colonies of naïve ESCs (Fig. 1A, bright-field images). Immunofluorescence analysis confirmed decreased expression of Nanog, a key pluripotent marker, in EpiSCs compared to ESCs (Fig. 1A, IF image). To further confirm the transition to a primed pluripotent state, RT-qPCR analysis for key marker genes was performed. Compared with ESCs, EpiSCs showed decreased expression of naïve markers (Oct4, Sox2, Nanog, Rex1, Esrrb) and increased expression of post-implantation epithelial cell markers (Sox17, Fgf5, Otx2) (Endoh & Niwa, 2022) (Fig. 1B). These results confirm that EpiSCs were successfully derived from BVSC ESCs and acquired morphological and molecular characteristics of primed state.
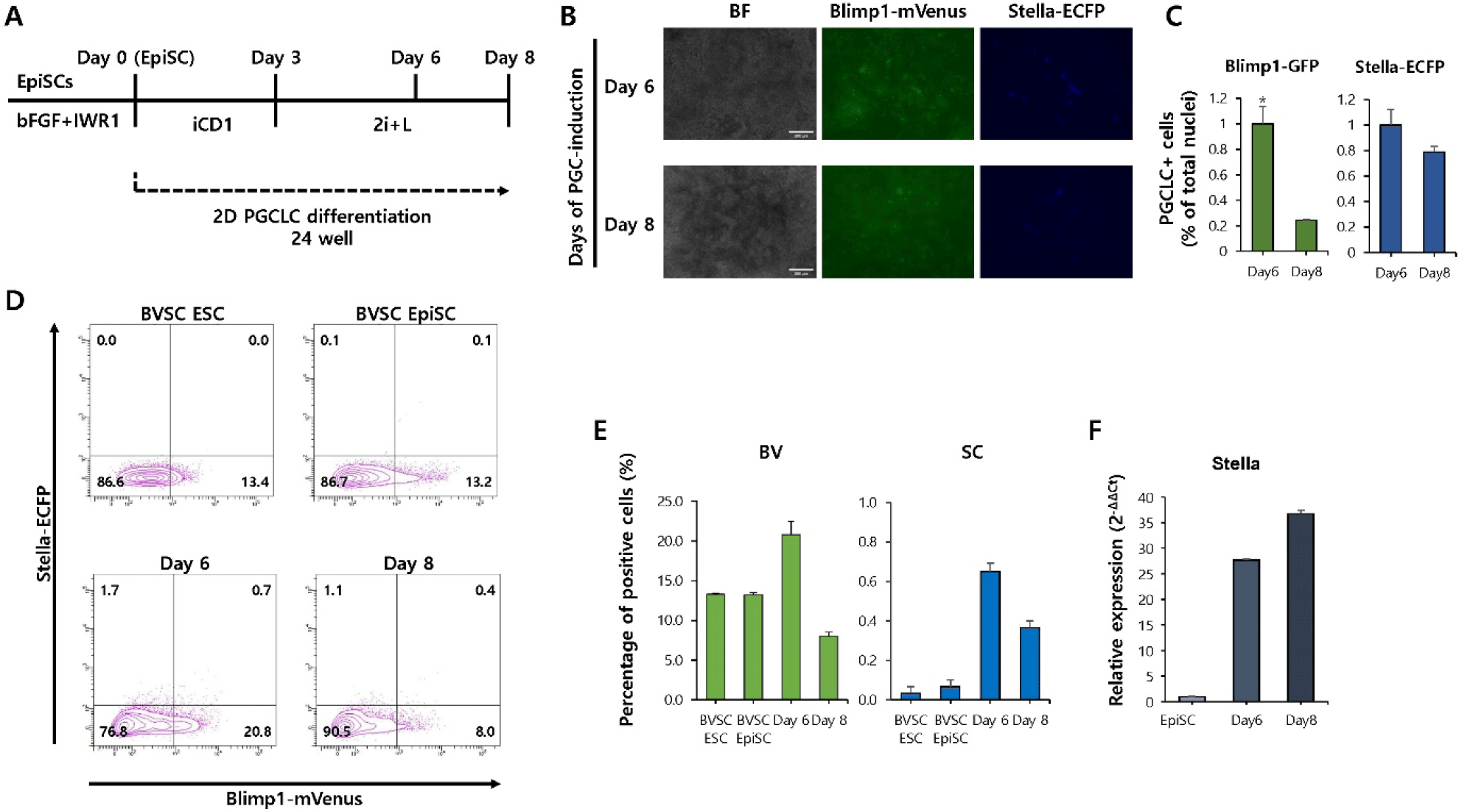
Traditionally, BMP4, stem cell factor (SCF), epidermal growth factor (EGF), and leukemia inhibitory factor (LIF) treatment were required to induce PGCLC from EpiLC. However, in the newly applied EpiSCs treatment method, PGCLC differentiation was successfully achieved by treating EpiSCs with BMP4, DOT1L inhibitor, and EZH2 inhibitor for 3 days, followed by culturing under 2i (MEK/ERK and GSK3 inhibitors)+LIF conditions for more than 6 days (Yu et al., 2021) (Fig. 2A). The efficiency of PGCLC induction was assessed by monitoring the expression of the reporter BVSC. Fluorescence microscopy showed that mVenus+cells appeared on day 6 and 8, but not ECFP+cells (Fig. 2B). To further quantify PGCLC induction efficiency, nuclear counterstaining was performed, and the proportion of mVenus- and Stella-positive cells was calculated relative to the total number of nuclei (Fig. 2C). Flow cytometry analysis also showed that the ratio of Blimp1-mVenus+/Stella-ECFP+double-positive cells remained low at both time points, and there was no significant increase from day 6 (0.7%) to day 8 (0.4%) (Fig. 2D and E). Furthermore, quantitative PCR analysis revealed a progressive increase in the expression of Stella, a key PGC marker, from day 6 to day 8 of differentiation (Fig. 2F). These results suggest that PGCLC induction from EpiSCs under 2D culture conditions is largely ineffective in EpiSCs.
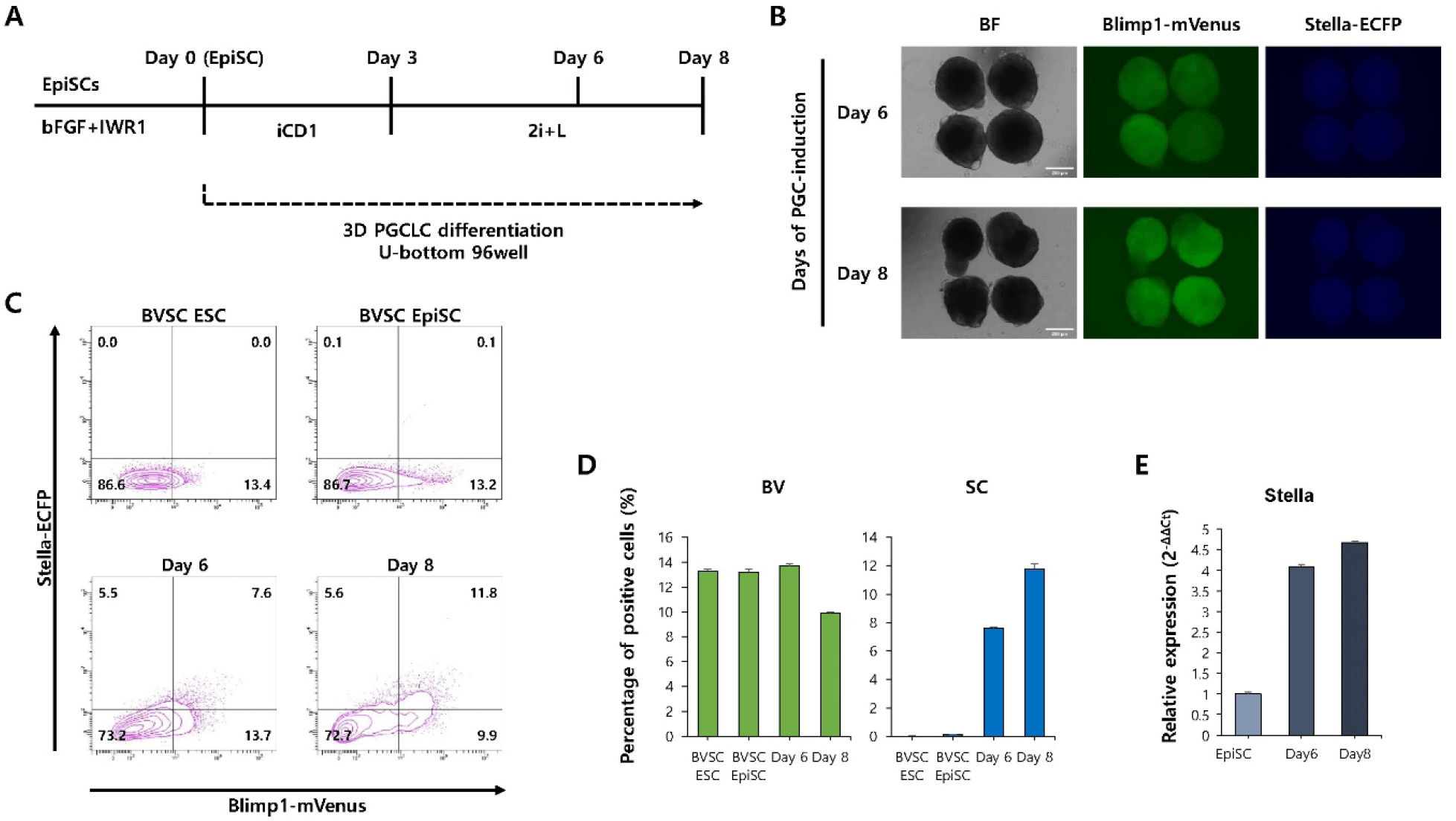
To evaluate whether spatial organization enhances PGCLC induction from EpiSCs, a 3D aggregation-based differentiation protocol was used (Fig. 3A). Fluorescence microscopy revealed a marked increase in the number and intensity of mVenus+cells and ECFP+cells within the aggregates at both day 6 and 8 (Fig. 3B). Flow cytometry analysis revealed a significant increase in the percentage of BVSC double-positive cells compared to 2D culture (Fig. 3C and D). In particular, the percentage of BVSC double-positive cells increased from day 6 (7.5%) to day 8 (11.8%), indicating that 3D culture conditions promote more robust and sustained PGCLC induction from EpiSCs. Consistently, quantitative PCR analysis revealed a progressive increase in the expression of Stella from day 6 to day 8 of differentiation (Fig. 3E).
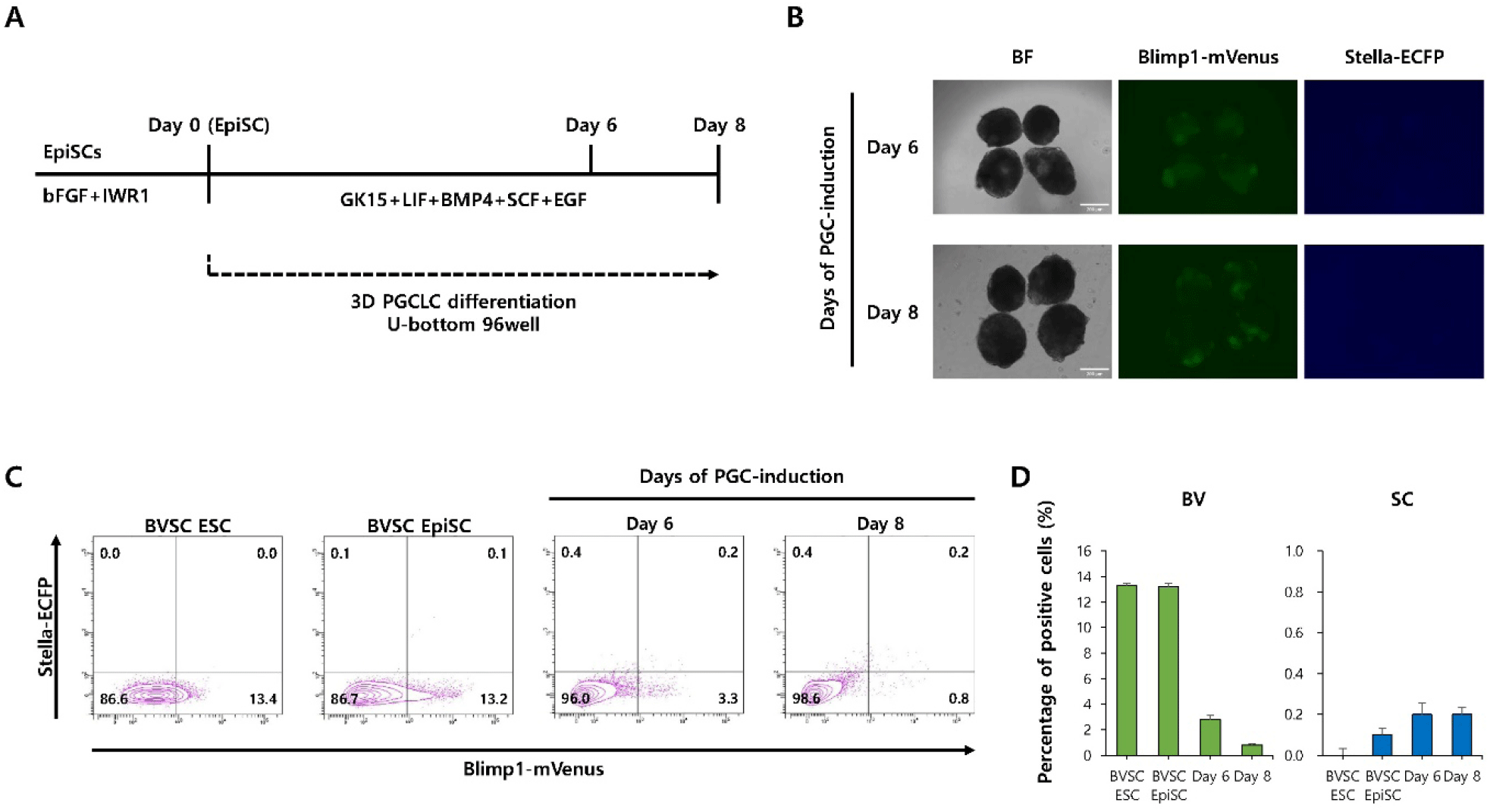
We next examined whether culture conditions optimized for EpiLC-derived PGCLC induction could also enhance PGCLC differentiation from EpiSC under 3D culture conditions (Fig. 4A). Fluorescence microscopy revealed the presence of mVenus+cells but not ECFP+cells (Fig. 4B). Flow cytometry further supported these observations, showing that the percentage of BVSC double-positive cells was relatively low at both time points (day 6: 0.2%, day 8: 0.2%) (Fig. 4C and D). These results suggest that PGCLC induction from EpiSCs is not effectively supported by EpiLC-optimized conditions, even under 3D culture environments.
DISCUSSION
EpiSCs are known to exhibit a stable primed pluripotent state that corresponds to the post-implantation epiblast stage (E5.5–7.5). Unlike naïve ESC-derived EpiLCs, EpiSCs have been reported to show a low efficiency of differentiation into PGCLCs, which is attributed to their reduced genetic stability and decreased responsiveness to germ cell-inducing signals such as BMP4 (Hayashi et al., 2011). As a result, most studies have focused on EpiLC-based PGCLC induction, while EpiSC-based studies are still limited (Hayashi et al., 2011; Ohta et al., 2017; Ooi et al., 2021; Li et al., 2025). Nevertheless, EpiSCs are an important model for post-implantation development, providing a unique platform to explore the molecular mechanisms of germ cell fate determination during this period.
In vitro differentiation is highly sensitive to the dimensions of the culture environment, which significantly influences cell fate. 2D monolayer culture systems are easy to manipulate and image, but they lack the spatial complexity and cell-to-cell interactions of in vivo tissues. In contrast, 3D aggregate cultures better mimic the structural features and signaling gradients of the embryo, providing a favorable environment for lineage-specific differentiation (Antoni et al., 2015; Urzì et al., 2023). Based on this, we hypothesized that a 3D aggregate environment could improve the efficiency of PGCLC induction from mouse EpiSCs.
Consistent with this hypothesis, PGCLC induction from mouse EpiSCs was significantly enhanced in a 3D aggregate environment compared to 2D monolayer culture. Flow cytometric analysis using BVSC-transgenic EpiSCs showed that the proportion of Blimp1-mVenus+/Stella-ECFP+ double-positive cells was significantly increased in 3D conditions, suggesting that spatial organization plays an important role in PGCLC induction. This enhanced effect may result from the formation of morphogenetic gradients (e.g., BMP4) and local signal amplification (e.g., Wnt/Nodal pathway) promoted by cell-cell interactions within the 3D aggregate (Metzger et al., 2017; Garnique et al., 2024).
Despite successful PGCLC induction from EpiSCs under 3D aggregate culture, our study also revealed that the culture media originally optimized for EpiLC-to-PGCLC differentiation failed to induce efficient PGCLC specification from EpiSCs, even under 3D conditions (Hayashi et al., 2011). This suggests that the limited differentiation potential is also influenced by intrinsic molecular and epigenetic differences between EpiLCs and EpiSCs (Hackett & Surani, 2014). EpiLCs are transient and unstable intermediates known to maintain a chromatin state suitable for germ cell induction, whereas EpiSCs are more advanced in development and exhibit characteristics of lineage differentiation readiness and epigenetic constraints (Factor et al., 2014). Therefore, signaling inputs designed for EpiLCs (e.g., BMP4, LIF, SCF, EGF) may not be sufficient to overcome the repressive transcriptional environment present in EpiSCs. Furthermore, the reduced responsiveness of EpiSCs to WNT and BMP signals may be due to reduced pathway sensitivity or silencing of key PGC-specific genes (e.g., Blimp1, Prdm14) (Ohinata et al., 2005), which could explain why EpiLC-based media fail in this context. These results highlight the need to establish tailored differentiation protocols that reflect the unique signaling and epigenetic environments of each pluripotent stem cell state. Additional studies comparing EpiLCs and EpiSCs during PGCLC induction, including phosphorylation signaling analysis, ATAC-seq, and single-cell RNA-seq, will provide valuable insights into the regulatory barriers that limit germline differentiation from more advanced pluripotent stem cells.
However, the present study did not directly evaluate the biological identity or functional potential of the induced PGCLCs. It remains unclear whether they undergo complete epigenetic reprogramming and can further colonize the gonad and develop into functional germ cells. Future studies should include transcriptome profiling, DNA methylation analysis, and transplantation assays to comprehensively assess the identity and maturation of 3D-derived PGCLCs. In addition, variability among different EpiSC lines was not addressed, which may impact the scalability and applicability of the protocol. Therefore, further studies should include transcriptome and DNA methylation profiling, transplantation assays, and validation across multiple EpiSC lines. Furthermore, when the PGCLC-inducing conditions optimized for EpiLCs were directly applied to EpiSCs, differentiation did not occur. This is likely due to mismatches in developmental stages and differences in signal responsiveness.
In summary, this study demonstrate that 3D culture conditions significantly enhance PGCLC induction from mouse EpiSCs compared to 2D culture systems. This suggests that 3D aggregates provide a useful platform to study germline fate determination and to refine differentiation protocols for developmental and regenerative medicine research.
