INTRODUCTION
Melatonin mediates environmental cues such as light and photoperiod and consequently influences important physiological processes including reproduction in mammals (Malpaux et al., 2001; Pevet, 2003) and in teleosts (Mayer et al., 1997; Falcón et al., 2007). However, the role of melatonin in the process of teleost reproduction seems less clear due to the descrepancies observed in the previous studies. In European eel, Anguilla anguilla, the effect of melatonin treatment on reproduction was inhibitory (Sébert et al., 2008) while the same treatments were stimulatory in carp, Cyprinus catla (Bhattacharya et al., 2007), seabass, Dicentrarchus labrax (Bayarri et al., 2004) and zebrafish, Danio rerio (Carnevali et al., 2011). Even within the same species, the effect of melatonin administration was dependent on gonadal stage or sex of subjects (Bhattacharya et al., 2007). This indicates that the action of melatonin on fish reproduction is far more complicated than we thought.
The effects of melatonin within the body of animals are known to be mediated through melatonin receptors that belong to the G protein-coupled receptor (GPCR) superfamily (Dubocovich, 1995; Reppert et al., 1996). Supporting this, in one of teleost fish, seabass, the effects of melatonin rely not only on its rhythmic production, but also on the rhythmic expression of its receptors (Bayarri et al., 2010). There are three different subtypes of melatonin receptors, mel 1a, mel 1b and mel 1c. While the mel 1a and mel 1b subtypes are found in all vertebrates investigated, the mel 1c subtype has been found only in non-mammalian so far (Ebisawa et al., 1994; Reppert et al., 1995; Wiechmann et al., 1999). However, which of these are specifically involved in fish reproduction including puberty is not known.
Puberty is the developmental period when an individual matures sexually for the first time, and activation of brain-pituitary-gonadal axis (BPG axis) is prerequisite for puberty. This activation of BPG axis is initiated by an increase of gonadotropin-releasing hormone (GnRH). GnRH released from hypothalamus promotes the maturation of gonad by inducing release of luteinizing hormone (LH) and follicle-stimulating hormone (FSH) via its cognate receptor located in pituitary gonadotropic cells. GnRHs are key regulators in sexual development and reproduction in vertebrates and there are two or three piscine GnRH genes encoding GnRH peptides of different structures (Parhar, 2002). Among three types of GnRH identified in Nile tilapia Oreochromis niloticus (Parhar et al., 2003), GnRH I is located in the preoptic area (POA) and pituitary showing a close relationship with the activation of BPG axis (White et al., 1995; Parhar et al., 1996; Gothilf et al., 1996). Thus, the expression of GnRH receptor I (GnRHr I) gene could be indicative of the activation of BPG axis in this species.
In this study, we cloned and analyzed the sequence of melatonin receptor 1a gene in Nile tilapia and examined the tissue distribution of three receptors, mel 1a, mel 1b and mel lc genes. Finally, we investigated which receptor is involved in the onset of puberty by comparing their gene expression with GnRHr I gene expression using quantitative real-time PCR.
MATERIALS AND METHODS
Mature male Nile tilapia (O. niloticus) was anaesthetized by benzocaine (50 ppm), and the whole brain was removed (body length: 16.2 cm, body weight: 70.5 g). The brain was homogenized with TRI REAGENT® (Molecular Research Center, Inc., Cincinnati, USA) to extract total RNA. Before RACE PCR, partial sequence of mel 1a was extended by RT-PCR using gene specific primers (Table 1) based on the mel 1a sequence of a close species: O. mossambicus (GenBank accession No. FJ531797.1). cDNA was synthesized from 2 μg of the total RNA with M-MLV reverse transcriptase (Promega, Madison, USA) and oligo d(T)15 primer (Promega, Madison, USA). PCR was carried out in 25 μl reaction mixture containing 1 μl cDNA, 400 nM forward and reverse primers (Table 1), 12.5 μl Go Taq Green Master Mix (Promega, Madison, USA) and nuclease-free water up to 25 μl. RT-PCR product was sequenced, and consequently the partial sequence (492 bp) was expanded to 858 bp. Based on the expanded mel 1a sequence, two forward primers and three reverse primers were designed (Table 1). RACE PCR was conducted using nTaq DNA Polymerase (Enzynomics™, Daejeon, Korea) and 5'/3' RACE Kit, 2nd Generation (Roche, Mannheim, Germany) according to the protocol. RACE PCR products were cloned using TOPcloner™ TA kit and Chemically Competent E. coli (Enzynomics™, Daejeon, Korea). Next, plasmid DNA was extracted and sequenced to find out the full sequence of mel 1a including 5' and 3'-UTR. The obtained mel 1a mRNA sequence was translated by ExPASy Translate Tool (http://www.expasy.ch/tools/dan.html) and the amino acid sequence was used to conserved domains analysis (http://www. ebi.ac.uk/Tools/InterProScan/). Mel 1a amino acid sequences from various species were obtained from GenBank (http://www.ncbi.nlm.nih.gov/) and aligned by ClustalW Multiple Alignment program and trimmed. A phylogenetic tree was formed using MEGA5 (Tamura et al., 2011) and Neighbor-Joining method (Saitou and Nei, 1987).
To investigate the tissue specific expressions of mel 1a, mel 1b and mel 1c, total RNA was isolated from whole brain, liver, gill, ovary, muscle, eye, heart, intestine, spleen and kidney of an adult female tilapia (body length: 15.0 cm, body weight: 57.9 g) and subjected to RT-PCR using primers for mel 1a, mel 1b and mel 1c genes (Table 2). PCR step was a cycle of preheating at 94°C for 2 min and 35 cycles of amplification (for mel 1a, mel 1b and mel 1c genes) or 25 cycles (for β-actin gene) including denaturation at 94°C for 30 sec, annealing (at 53°C for mel 1a, 57°C for mel 1b, 53°C for mel 1c or 55°C for β-actin gene) for 1 min and extension at 72°C for 1 min, followed by a cycle of final extension at 72°C for 5 min. The PCR products were electrophoresed on 1% agarose gels and stained with ethidium bromide. The products were, then, visualized under UV light and analyzed by Image Analysis System (Kodak DC290).
| Sequence | Product size (bp) | ||
|---|---|---|---|
| Tissue distribution (RT-PCR) | mel 1a F | 5'-ACG CAG GGA ACA TCT TTG TG-3' | 318 |
| mel 1a R | 5'-CAC AAA CAG GTT GGG CAC GAT-3' | ||
| mel 1b F | 5'-TCG CCC AGA ATG TCA GCA GT-3' | 221 | |
| mel 1b R | 5'-GGC CCA GCA AAT GGC AAA CA-3' | ||
| mel 1c F | 5'-TAC CCC TAC CCA CTG GTC CT-3' | 234 | |
| mel 1c R | 5'-TGG CGG TGA GTA GCC AAG TA-3' | ||
| β-actin F | 5'-AAT CGT GCG TGA CAT CAA GG-3' | 392 | |
| β-actin R | 5'-AGT ATT TAC GCT CAG GTG GG-3' | ||
| Quantitative real-time PCR | qmel 1a F | 5'-GGT TGT GAT TCC CCT CAT CCC-3' | 105 |
| qmel 1a R | 5'-TTC TGG TTC AGC ACG CCA TAC A-3' | ||
| GnRHr I F * | 5'-GTG GCT TGC CGG AGA CTT TG-3' | 123 | |
| GnRHr I R * | 5'-AGA GGG TTG AGG ATG GCT GAC T-3' | ||
| qβ-actin F | 5'-AGC ATC CCG TCC TGC TCA CA-3' | 121 | |
| qβ-actin R | 5'-AGC ACA GCC TGG ATG GCA AC-3' | ||
GnRHr I gene primer from Martinez-Chavez et al. (2008). Primers for mel 1a, mel 1b (GenBank accession No. XM003446809.1), mel 1c (Kim et al., unpublished) and β-actin (GenBank accession No. XM003443127.1) genes were newly designed.
Experimental fish were produced from Nile tilapia obtained from Chungju, Chungcheongbuk-do in Korea. They were reared in a closed recirculating aquaculture system at 27 ± 1°C under natural photoperiod (sampling period: July to December) and fed twice a day. Fish from a batch were sampled 1 to 4 week intervals at noon (12 pm) from 1 week post-hatch (wph) to 24 wph. Depending on the growth of the fish, their heads (1—7 wph) or brains (10—24 wph) were sampled (n=4). Protocols of total RNA extraction and cDNA synthesis are the same as above. We examined the expression of GnRHr I and mel 1a genes together with the expression of β-actin gene for normalization. Quantitative real-time PCR reactions were conducted using QuantiMix SYBR (PhileKorea, Seoul, Korea) and Eco™ Real-Time PCR System (illumina®, San Diego, USA) in a 20 μl reaction mixture containing 1 μl cDNA, 10 μl QuantiMix SYBR, 250 nM forward and reverse primers (Table 2) and nuclease-free water up to 20 μl. Melting curve analysis was also included at the end of the PCR. Each expression value of the mRNAs was presented as Ct (threshold cycles) values. The levels of the mRNAs were normalized against the amount of β-actin mRNA and the relative level was determined by the comparative threshold cycle method, 2-ΔCt.
Data were presented as mean ± S.E.M. For statistical analysis of mel 1a and GnRHr I mRNA expression, expression level observed at each wph was compared with the level observed immediately previous wph within this study by independent t-test (p<0.05) using SPSS version 18.0 for windows.
RESULTS
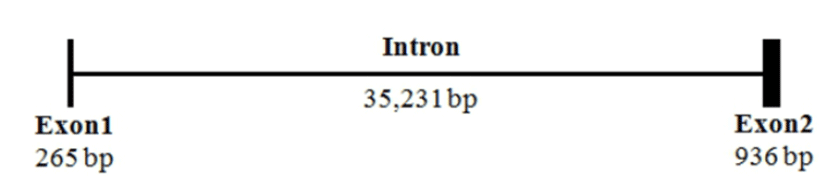
The sequence of mel 1a mRNA cloned from the brain of Nile tilapia consisted of an ORF (1053 nt), a 82 nt 5'-UTR and a 49 nt 3'-UTR (Fig. 1). Deduced amino acid sequence of mel 1a contained a signal peptide region, predicted transmembrane domains and conserved N-R-Y motif. By comparison of mel 1a mRNA sequence with that of genome sequence in this species (GenBank accession No. NT167456), mel 1a gene of O. niloticus was found to be comprised of two relatively short exons (265 and 936 bp, respectively) and one bulky intron of 35,231 bp between the two exons (Fig. 2).

Multiple alignment of Nile tilapia mel 1a amino acid sequence with those of other fish species and subsequent phylogenetic analysis (Fig. 3) showed high similarity of this gene with that of other fish species such as Mozambique tilapia (O. mossambicus, 99%), European seabass (D. labrax, 97%), rabbitfish (Siganus guttatus, 96%) and grass puffer (Takifugu niphobles, 96%).
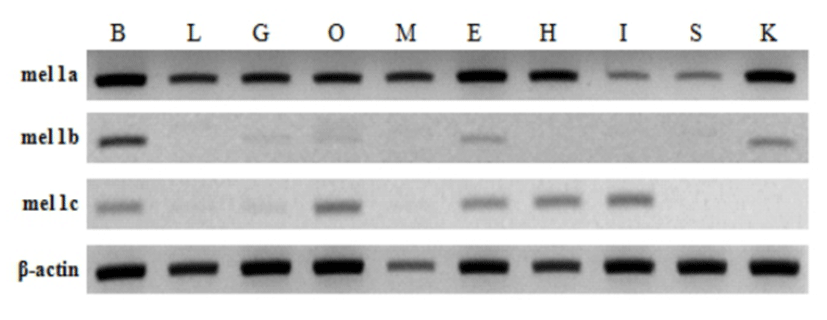
Tissue specific expression of melatonin receptors analyzed by RT-PCR was shown in Fig. 4. Mel 1a gene was expressed abundantly and ubiquitously in all tissues investigated including the brain and various peripheral tissues: liver, gill, ovary, muscle, eye, heart, intestine, spleen and kidney. In contrast, mel 1b and mel 1c genes were faintly detected in only several tissues. Mel 1b transcript existed in the brain, gill, ovary, eye and kidney, and mel 1c transcript was distributed in the brain, gill, ovary, eye, heart and intestine. Three different melatonin receptors were commonly detected in the brain, gill, ovary and eye.
The relative expression level of mel 1a gene significantly increased at 4 wph and decreased at 7 wph compared with the respective previous investigation. Then it remained low until 12 wph and increased significantly at 13 wph and 14 wph compared with the previous investigation (Fig. 5A). The relative expression level constantly increased until 15 wph and reached the highest level though not significantly different from the level at 14 wph. Then, it declined rapidly at 16 wph and 20 wph. On the whole, the level of mel 1a transcript exhibited a prominent increase between 13—16 wph.
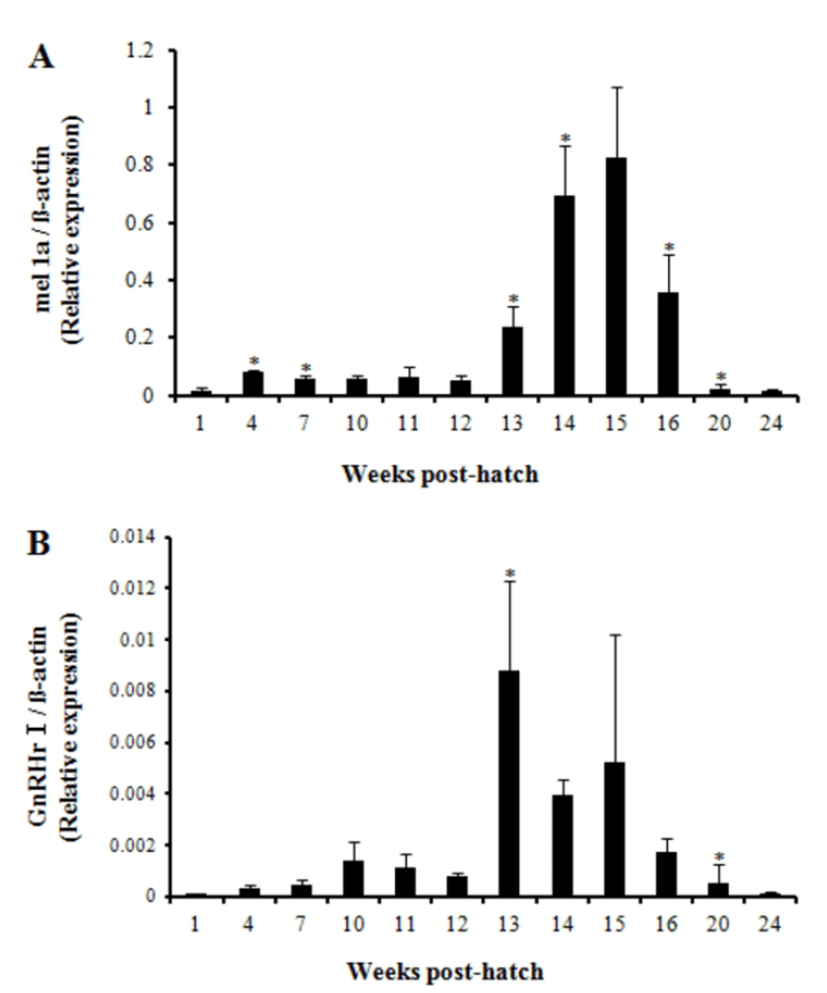
The first significant increase in the level of relative GnRHr I gene expression was observed at 13 wph (Fig. 5B). Then, the expression level maintained up to 15 wph though it tended to be decreased. Overall, expression profile of mel 1a gene that exhibited higher expression between 13—16 wph approximately corresponded to the profile of GnRHr I gene.
DISCUSSIONS
Mel 1a gene cloned from Nile tilapia was a rhodopsin like GPCR and had 7 transmembrane domains. While most of rhodopsin-like GPCRs have a consensus sequence of D/E-R-Y at the end of third transmembrane domain, melatonin receptors have a conserved sequence of N-R-Y at the same location (Kokkola et al., 2005). Anterior two residues of N-R-Y motif are known to be involved directly in the activation of rhodopsin and β-adrenergic receptors (Ballesteros et al., 2001; Gether et al., 2002). The mel 1a gene structure of O. niloticus that had two exons and one bulky intron (35 kilobases) was similar to that of mouse (Roca et al., 1996). In addition, the mel 1a gene in tilapia showed high similarity to the same gene from different teleost species. These data indicates that mel 1a is highly conserved gene.
The coding sequence (CDS) of mel 1a gene identified in this study was 100% identical to CDS of predicted O. niloticus melatonin receptor subtype 1A-A-like (GenBank accession No. XM003449697) predicted from the genomic sequence (NT167456) using GNOMON method. The presence of 1A-like sequence (GenBank accession No. XM003447612.1) distinguished from 1A-A-like suggests the possible existence of two subtypes of mel 1a gene in this species. It is necessary to confirm this possibility by further studies. So far, teleost species which have two subtypes of mel 1a are zebrafish (Reppert et al., 1995), rainbow trout (Mazurais et al., 1999) and goldfish (Ikegami et al., 2009).
All three types of Nile tilapia melatonin receptor genes were expressed in the brain, eye and ovary in common. Especially, they were abundant in the brain and eye. Even though these receptors were expressed abundantly in the brain and eye in common, mel 1a mRNA was the most abundant suggesting that this receptor mediate most of melatonin actions at the level of central nervous system. Mel 1a gene was also prominently expressed in all peripheral tissues investigated in this study, including liver, gill, ovary, muscle, heart, intestine, spleen and kidney while mel 1b and mel 1c genes were expressed in fewer tissues. The functional significance of peripheral melatonin receptors mediating the melatonin effects needs further investigation.
In this study, expression of GnRHr I gene was used to verify the timing of puberty onset in Nile tilapia, and it was compared with the expression pattern of mel 1a gene since the increase of GnRHr I gene expression during puberty had been frequently reported in fish (Mohamed et al., 2007; Martinez-Chavez et al., 2008; Park et al., 2012). Martinez-Chavez et al. (2008) reported 9—13 wph as the timing of puberty onset in Nile tilapia by putting together the results of gonadal histological observation and the significant expression of GnRHr I. In the present study, the first significant rise of GnRHr I gene expression occurred at 13 wph, implying the timing of puberty onset. Difference in the timing of puberty onset between these two studies may have resulted from different rearing conditions, strains and growth rates between two experimental systems (Park et al., 2012).
Relative expression of both mel 1a and GnRHr I gene increased significantly at 13 wph, the putative timing of puberty. This coinciding elevation of both gene expression indicates melatonin/mel 1a system may have a role in puberty onset through GnRH system. In male masu salmon Oncorhynchus masou pituitary, GnRH and LH levels were decreased by melatonin treatment, resulting the decrease of gonado-somatic index (GSI) and plasma T level (Amano et al., 2004). Similarly, melatonin administration in A. anguilla partly inhibited gonadal development by stimulating dopaminergic inhibitory actions on pituitary-gonad axis (Sébert et al., 2008). In zebrafish, however, melatonin administration via water led to a significant increase of GnRH III gene expression in the brain and LH gene expression in the pituitary resulting the increase of GSI (Carnevali et al., 2011). In addition, numerous melatonin-binding sites were distributed in POA where GnRH and dopamine neurons were located in various teleost species (reviewed by Dufour et al., 2010) and in tilapia hybrid (O. aureus × O. niloticus) (Levavi-Sivan et al., 2004). Despite of the discrepancies between studies, all of these data commonly indicate that melatonin would have negative or positive effects on reproduction via GnRH/dopamine system in teleost.
The concomitant increase of both GnRHr I and mel 1a gene expression suggests the possible involvement of melatonin in the onset of puberty via mel 1a. In support of this, in male seabass, the plasma melatonin level increased during the first sexual maturation (Bayarri et al., 2010). In Senegalese sole Solea senegalensis exhibiting annual reproductive rhythms, a seasonal variation of mel 1a gene expression in pituitary was observed, showing an increase after main spawning season and decrease in second spawning period (Confente et al., 2010). Changes of plasma melatonin level positively corresponded to the seasonal variation in the same species (Vera et al., 2007).
In summary, we have cloned melatonin receptor 1a gene from the brain of Nile tilapia. This gene was highly conserved among species based on the phylogenetic and sequence analysis. Mel 1a mRNA was most abundant in all tissues investigated in this study, suggesting that the considerable melatonin actions are mediated by mel 1a in these peripheral tissues. Gene expression profiles of GnRHr I revealed a putative timing of puberty onset at 13 wph, and expression of mel 1a gene showed a close relationship with puberty onset in this species. Further studies should clarify the exact function of mel 1a gene that was concomitantly increased with GnRHr I gene during the putative puberty onset.