INTRODUCTION
The olive flounder (Paralichthys olivaceus), is widely distributed along the coast of East Asia and is one of the most economically important marine species in the region. P. olivaceus is successfully cultured in Japan, China and Korea. But, olive flounder faces significant mortality especially in the early stages of life, which attracted our interest to characterize how their immune system works during the development stages. In fishes, larvae are immediately exposed to various microorganisms after hatching (Zapata et al., 1997). However, the immune system is still developing and not all of the structures and functions present in the adults are in the larvae (Ellis, 1988; Tatner, 1996). Therefore, knowledge of the immune system in ontogeny can offer new protective strategies and indicate optimal vaccination point (Vadstein, 1997).
Previous studies show that the appearance of the immune organ and system during early developmental stages is different in different fish species, such as sea bream (Jósefsson & Tatner, 1993), turbot (Padrós & Crespo, 1996), yellowtail and olive flounder (Chantanachookhin et al., 1991). Most of these studies have been carried out using histology as the main technique to investigate the development of the immune organs in fish.
Recently, lymphoid organs and both T and B cells have been chosen to study the ontogeny of the fish immune system in various fish species, and recombination-activating gene 1 (RAG-1) and immunoglobulin M (IgM) have been considered as very useful markers of the immunological maturation (Corripio-Miyar et al., 2007; Hansen & Kaattari, 1995; Willett et al., 1997; Peixoto et al., 2000; Huttenhuis et al., 2005; Zhang et al., 2009; Fan et al., 2009).
The RAG-1 gene is essential in the differentiation of immature B and T cells and is expressed in primary lymphoid organs, although not in mature cells (Nagaoka et al., 2000). It is this expression during the maturation of the lymphoid organs that makes the RAG-1 gene such a useful marker for the study of the development of these organs (Willett et al., 1997; Dunham, 1999). IgM is also an important gene that can be used in the study of the ontogenesis of the immune system, as it is the first formed antibody of the primary response in higher vertebrates (Magnadóttur, 1998). The presence of IgM has been extensively reported in many fish, including studies of its appearance during development and even maternal transfer of IgM to the offspring (Kanlis et al., 1995; Olsen & Press, 1997; Takemura & Takano, 1997; Picchietti et al., 2001). In teleost fish, RAG-1 and IgM has been studied in gadoid haddock (Corripio-Miyar et al., 2007), zebra fish (Willett et al., 1997), common carp (Huttenhuis et al., 2005), redspotted grouper (Mao et al., 2012) and striped trumpeter (Covello et al., 2013).
The present study aimed to improve knowledge about expression of RAG-1 and IgM in olive flounder, which can provide crucial information regarding the ontogeny of the immune system, with implications for vaccination regimes. Although the RAG-1 and IgM gene has been reported in several fish, considerably less is known about the expression of both genes in olive flounder. Therefore, we investigate the expression pattern analysis in larvae and tissue-specific expression in immune organ.
MATERIALS AND METHODS
Olive flounder larvae was obtained from Genetics and Breeding Research Center, National Fisheries Research and Development Institute (NFRDI; Geoje, Republic of Korea), and maintained in 10 tons flow through tank at 20±1°C under a natural photoperiod. The sample was prepared from various tissues including brain, muscle, fin, eye, liver, gill, kidney, and spleen obtained from 90-day-old healthy olive flounder (n=3). Different larval stages including 5 to 55 days post hatching (dph) (5 dph, 10 dph, 15 dph, 20 dph, 25 dph, 30 dph, 35 dph, 40 dph, 45 dph, 50 dph and 55 dph) fish and unfertilized oocyte kept at 18.0°C in the sea-water tank. The sample of 10 randomly selected fishes were collected and immediately frozen in liquid nitrogen, and stored at –80°C until RNA extraction.
Total RNA was isolated with Trizol Reagent (Invitrogen), following the manufacturer’s instructions. RNA was suspended in DEPC-treated water and used for RT-PCR. The total RNA concentration was quantified by spectrophotometer and 1μg of total RNA was used for reverse transcribed into cDNA using First Strand cDNA synthesis kit (Roche). The amplification was performed with AmpliTag Gold DNA Polymerase (Applied Biosystems) in thermal Cycler (Applied Biosystems) using the following parameters: denaturation at 95°C for 10 min and 35 cycles of reactions of denaturation at 98°C for 10 sec, annealing at 58°C for 30 sec, and elongation at 72°C for 30 sec. An aliquot of each PCR product was subjected to 1.2% (w/v) agarose gel electrophoresis and visualized by staining with ethidium bromide.
All the primers were designed using Primer3 (Rozen & Skaletsky 2000). The gene specific primers are listed in Table 1.
To evaluate mRNA levels of RAG-1 and IgM, primers were specifically designed to detect and quantify cDNA sequences without detecting genomic DNA. The real-time PCR reactions were monitored with melting curve analysis using 7500 software (version 2.0.5). Amplification efficiency was determined by a serial dilutions. All experiments were repeated in triplicate. Each reactions displayed an efficiency 95.9% (R2=0.994), 96.7% (R2=0.997) and 98% (R2=0.999), RAG-1, IgM and β-actin, respectively.
Real-time PCR reaction mixture contained 2 μL of diluted cDNA sample, specific primer and the FAST SYBR Green PCR Master Mix (Applied Biosystems). The reactions for the amplification of RAG-1, IgM and β-actin were subjected to an initial denaturation at 95°C for 20 sec, annealing at 58°C for 30 sec, and elongation at 60°C for 30 sec. Thermal cycling and fluorescence detection were conducted using the 7500 Fast Real-Time PCR system (Applied Biosystems).
Expression of RAG-1 and IgM gene was normalized to an endogenous reference β-actin and presented as subtraction of target CT values from β-actin CT values (ΔCT value). Comparison of gene expression between tissues and calibrator was derived from subtraction of the calibrator ΔCT values from the target ΔCT values to give a ΔΔCT value, and relative gene expression was calculated to determine fold difference (2-ΔΔCT).
The data are presented as the mean ± standard deviation. The mRNA levels of RAG-1 and IgM was expressed as a ratio to those β-actin, which was simultaneously amplified as an internal control for each cDNA. The data was statistically analyzed by one-way ANOVA after arcsine transformation when needed, and followed by a Tukey test for identification of the statistically distinct groups. Significant differences were accepted for P <0.05.
RESULTS
To investigate the expression of the RAG-1 and IgM genes, oligonucleotide specific primers were designed according to previously reported both genes in olive flounder (GenBank accession no. KC442210 and AB052744, respectively) (Table 1). In order to investigate of olive flounder RAG-1 and IgM mRNA expression in various tissues, we assessed the expression of the both genes in various tissues from 90-days-old healthy olive flounder by RT-PCR (Fig. 1). It was found that RAG-1 and IgM mRNAs are significantly expressed in the spleen, kidney and gill, not expressed in other tissues. These overall patterns of RAG-1 and IgM mRNA expression were limited in typical immune organs. Kidney, spleen and gill were typical immune organs in teleost fish. The kidney is a major organ with key regulatory functions for the immune-endocrine interactions, and spleen as a secondary lymphatic and scavenging organ plays a crucial role in hematopoiesis, antigen degradation and antibody production processing (Rauta et al., 2012). Also, gill is the main mucosal surface and immune barriers.
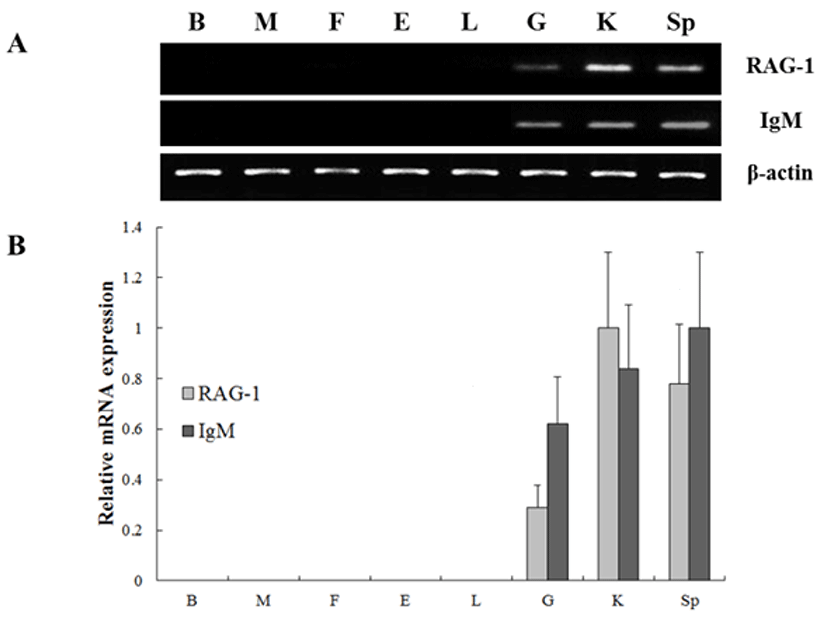
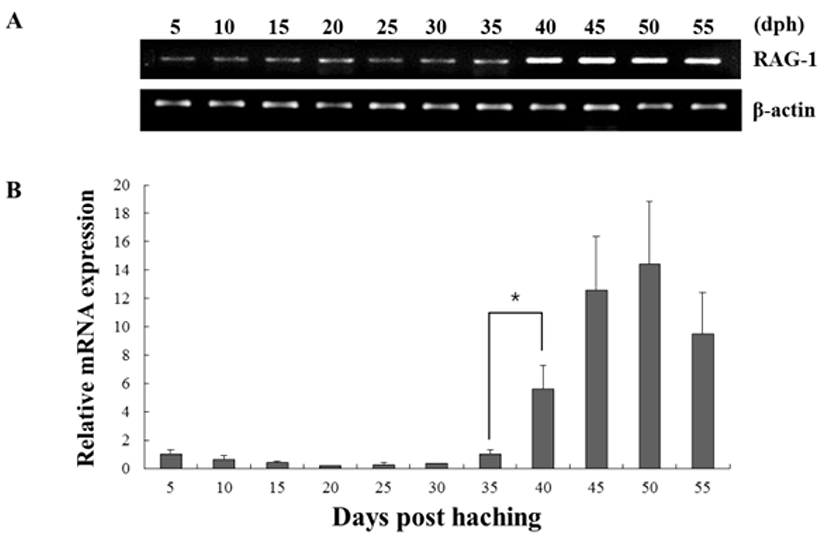
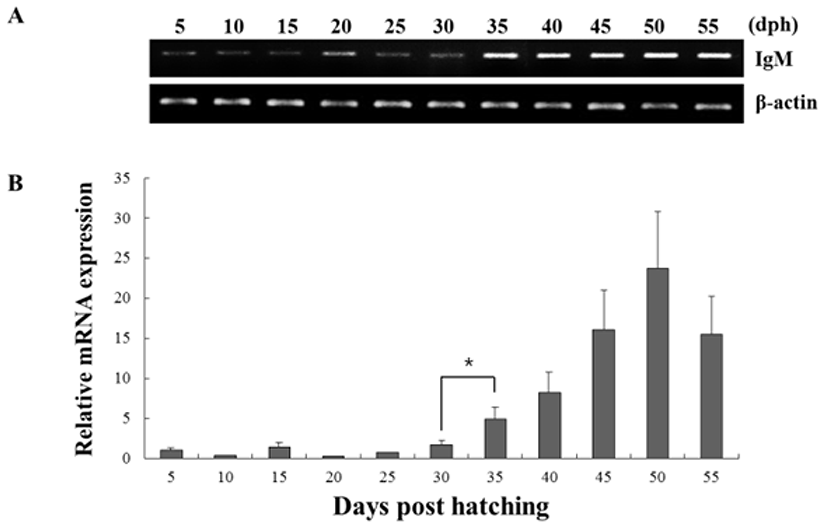
The expression of both genes from triplicate samples was standardized using housekeeping gene β-actin. The expression pattern of the RAG-1 and IgM gene was investigated over time course 5, 10, 15, 20, 25, 30, 35, 40, 45, 50 and 55 dph of olive flounder using real-time PCR. RAG-1 expression was first detected at 5 dph, and signifycant expression was observed in the 40, 45, 50 and 55 dph (Fig. 2). The intensity of RAG-1 transcript was measured on the basis of 5 dph (1.0-fold), each value has the 40 dph (5.6-fold), 45 dph (12.6-fold), 50 dph (14.5-fold) and 55 dph (9.5-fold), respectively. IgM mRNA transcripts were first detected at 5 dph, and the transcription level was increased obviously in the 35 dph (Fig. 3). The intensity of IgM transcript was measured on the basis of 5 dph (1.0-fold), each value has the 35 dph (4.9-fold), 40 dph (8.3-fold), 45 dph (16.1-fold), 50 dph (23.7-fold) and 55 dph (15.5-fold), respectively. As RAG-1 and IgM mRNA was detected earlier than expected (5 dph), the possibility of maternal mRNA transfer into oocytes was investigated. It was found that both RAG-1 and IgM mRNA transcripts were present in unfertilized oocytes (Fig. 4). Studies on several fish species have shown that RAG-1 and IgM are transferred from the mother to the offspring (Covello et al., 2013; Takemura & Takano, 1997), and may be involved in the early defense against pathogen in eggs containing developing embryo and larvae.

DISCUSSIONS
Because of the importance of immune response, many studies have been conducted in fish during the early larvae stages. This study is the first to report the mRNA expression of the important immune genes RAG-1 and IgM in the olive flounder. In addition their expression was followed from unfertilized oocytes through to the early developmental stage of the larvae. As both genes are important markers of the ontogeny of the adaptive immune response, their level of expression can help from the problem of high mortality in olive flounder larvae. In teleost fish, the RAG-1 mRNA level was first examined in lymphoid organs of rainbow trout larvae (Hansen & Kaattari, 1995), and also investigated in the same organs of zebra fish (Willett et al., 1997) and common carp (Huttenhuis et al., 2005). Humoral adaptive immunity in fish is mediated by Ig, and the IgM class is the primary immunoglobulin in most teleost fish. The IgM class has been studied in many species such as salmon (Hatten et al., 2001), zebra fish (Danilova et al., 2000), Atlantic cod (Schrøder et al., 1998) and rainbow trout (Lee et al., 1993).
In our study, RAG-1 and IgM mRNA was investigated in various tissues including brain, muscle, fin, eye, liver, gill, kidney, and spleen (Fig. 1). The results confirmed in both genes that the spleen, kidney and gill are the typically immune organs in teleost fish species, since the differrentiation of early T and B cells occur in the tissues. In previous study, IgM gene was isolated and identified from the cDNA clones in olive flounder BAC library, and confirmed expression levels of various tissues using RT-PCR (Hirono et al, 2003). The results are similar with our study, showed that IgM transcript level was detected in spleen and kidney. In contrast, these results are conflicted with IgM expression studies in the red-spotted grouper (Mao et al., 2012), where IgM expression appeared to be ubiquitous and constitutive, except the gonad.
Fish larvae are exposed to pathogens a long period before of time until maturation of their lymphoid organs, therefore information of immune gene may be essential for survival (Zapata et al., 1997). However, the mRNA expression analysis of RAG-1 and IgM during the early developmental stage in olive flounder is insufficient. In this study, to information of RAG-1 and IgM expression during early larvae stages of development, we analyzed the expression pattern of both genes after hatching. As a result, RAG-1 and IgM mRNA was first detected in 5 dph, observed the increased mostly until 50 dph (Fig. 2). Significant expression of RAG-1 was detected in 40 to 55 dph, while IgM was detected in 35 to 55 dph. In our study, the immune system of B and T cell marked by RAG-1 and IgM starts to develop its function around 5 dph in olive flounder. The patterns of RAG-1 expression levels, they did not follow the trend of an initial increase in IgM mRNA expression. Studies in striped trumpeter have shown a similar pattern of RAG-1 and IgM expression, but with some variation in the peak expression patterns (Covello et al., 2013). This early detection of IgM mRNA led to the investigation of transcript presence in unfertilized oocytes (Fig. 4). Both RAG-1 and IgM mRNA were present in oocytes, leading to the conclusion that they were maternally transferred.
In summary, this study provides the information of RAG-1 and IgM mRNA expression during the early developmental stage in olive flounder. These expression pattern of both genes in tissue distribution and developmental stage shows that may play an important function in the early immune response of olive flounder larvae. This study is the first to report the mRNA expression of the important immune genes RAG-1 and IgM in the olive flounder. This will allow the timing of vaccination of fish to be optimized so as to allow protection at the earliest time possible, to help reduce the high mortalities seen during the early stages of development.
