INTRODUCTION
The olive flounder (Paralichthys olivaceus) is an economically important fish in Korea and Japan, and total yields of aquacultured olive flounder increased and reached to 40,805 tons in 2011 (FAO) in Korea. Recently, breeding research is focused only culture techniques and development of research also insufficient to further increase production. Higher breeding research including the development of molecular markers is essential to produce of good breeds and high value fisheries and to enhance our competitiveness in fisheries industry. Identification and functional characterization of economic traits (or breeding traits) related genes, the development of molecular breeding techniques using DNA markers to molecular breeding is meaningful (Hubert et al., 2009; Yoon, 2006). Moreover, application of these genes in the aquaculture (molecular selective breeding) has attracted worldwide attention.
Recently, large-scale DNA sequencing such as BAC library projects and EST projects have provided usable data for a great variety of species (Hayes et al., 2005). In particular, the sequence analysis was significantly increased in economic worth of fish. This information can be used to increase the accuracy of selection breeding research for economical traits, thereby increasing the rate of genetic gain and production efficiency.
The molecular marker is an important element for breeding research, and there are various methods to used for the research. The kind of molecular techniques are RAPD (random-amplified polymorphic DNA), RFLP (restriction fragment length polymorphism), AFLP (amplified fragment length polymorphism), MS (microsatellites) and SNP (single nucleotide polymorphisms) (Liu & Cordes, 2004). Among possible molecular markers, microsatellites have been most useful for population genotyping technology, but is not suitable to high-quality data about influence gene expression and the functions. Unlike MS, single nucleotide polymerphisms (SNPs) are the most abundant type of variation in DNA sequence among individuals in a population (Fahrenkrug et al., 2002). Moreover, these markers appear to have low mutation rates (a mutation occurs every 2 × 108 nucleotides) and hence are extremely stable than other genetic markers (The International SNP Map Working Group, 2001). Although, SNPs have been recently proposed as an alternative to microsatellites for genotyping individuals (Vignal et al., 2002), have a disadvantage of time and cost-consuming when a certain selection mechanism does not exist. Whereas, if developed SNPs from of expressed sequence tags (ESTs) can be particularly valuable, also it is possible to reduce the time and cost for developing a useful SNP marker. In aquaculture species, SNPs are especially important if they cause differences in economic traits, or are linked to the mutations that do so.
For some aquaculture species, a large number of SNPs was developed from expressed sequence tag (EST) database as below; catfish (He et al., 2003), salmon (Hayes et al., 2007; Campbell & Narum, 2011; Everett et al., 2011), sea bream (Hayes et al., 2007; Campbell & Narum, 2011; Everett et al., 2011), Atlantic cod (Hubert, 2009), and turbot (Vera et al., 2011). But, SNP marker was hardly developed and validated in olive flounder even economically important fish species in Korea (He et al., 2011; He et al., 2012).
Wild and cultured olive flounder have different growth rate and resistance to disease because of differences in the growth environment. For example, wild olive flounder has generally small size and slow growth but strong immunity, whereas cultured have generally rapid growth and heavy weight but weak immunity. Each feature is associated with a SNP, and therefore SNP analysis using wild and culture samples can be a help to find regions involved superior traits. In this study, we develop useful marker to detect the single nucleotide polymorphism (SNP) from EST in olive flounder and then apply to sample of wild and cultured. Moreover, we were identified substitution of SNP and population diversity analysis using developed marker.
MATERIALS AND METHODS
In this study used polymorphic sites were identified from expressed sequence tags (ESTs) derived from P. olivaceus. Using to the previous studied EST data, bioinformatic analysis was conducted to determine gene identities using Gene Master Software (Ensoltek). ESTs were assembled in clusters of contiguous sequences (contig) using ICAtools program (Parsons, 1995). Gene annotation procedures and homology searches of the sequenced ESTs have been locally done by BLASTX for amino acid similarity comparisons. Matches with the Expect value (E) less than 10xe-4 were considered to be significant. All ESTs that were not identified as orthologs of known genes were designated as unknown EST clones.
Total of 755 SNP genes were selected from chromosome 24. The selection of SNP gene criteria is as below; the frequency of minor allele is 30 percent, each gene is related with immune and growth, and evenly exists on chromosome 24. As a result, 144 markers were selected, developed the primer for detecting of specific gene. Primers of SNPs were designed using Primer 3 software (Rozen and Skaletsky, 2000) adjusting length products to 100-300 bp (Table 1).
Wild and cultured each of eight olive flounder were randomly obtained from East Sea and Jeju Island in Korea, respectively. Genomic DNA was extracted from pectoral fin with the standard method of phenol-chloroform (He et al., 2008) and a total of 144 primers were designed.
Amplification was carried out in the following conditions; in 20 uL volume including 2~4 uL template DNA (100~ 150 ng), 1× Gold Taq buffer (Amplified Biosystems), 2.5 mM of MgCl2, 0.2 mM of dNTP (Solgent), 10 pmol of both forward and reverse primers, 1.25 U of Gold taq DNA polymerase (Amplified biosystems) and 1 uL dimethyl sulfoxide (Sigma). Thermal cycling was conducted on a Applied Biosystems (Model #: 9902) as following conditions: initial denaturation at 95°C for 2 min, 35 cycles of denaturation at 95°C for 30 s, annealing temperature at 58°C for 30 s, and extension at 72°C for 30 s. Final extension step was performed at 72°C for 10 min. Amplification of each SNP was checked on 2% agarose gel using electrophoresis. PCR product was purified using DNA purification Kit, sequenced in forward direction with the Big Dye termination reaction chemistry kit (Nucleogen), and analyzed with an ABI 3730xl Genetic Analyzer (Applied Biosystems).
Sequences were aligned with the CLUSTAL W algorithm using MEGA 5.05 software. Heterozygosity was estimated using Arlequin software 3.5 and polymorphic information content (PIC) value using Power Marker V.3.25 software.
RESULTS
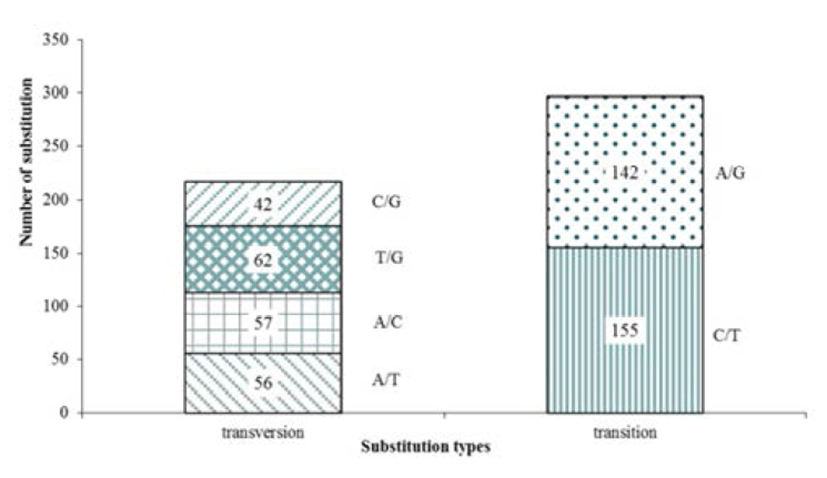
The olive flounder EST-derived database contains 4,327 ESTs and identified 693 contigs and 514 SNP were detected. The average of SNP frequency was 1 per 216 bp of contig sequences. These substitutions included 297 transitions (C/T, 155; A/G, 142) and 217 transversions (A/T, 56; A/C, 57; T/G, 62; C/G, 42), with C/T rate (155, 52.8%) has captured most of substitutions and C/G being the least common (42, 19.4%) alteration observed (Fig. 1). A transition/ transversion ratio (ts/tv) was 1.369, SNP substitutions result of development marker shows transitions rate higher than transversions rate (Table 2).
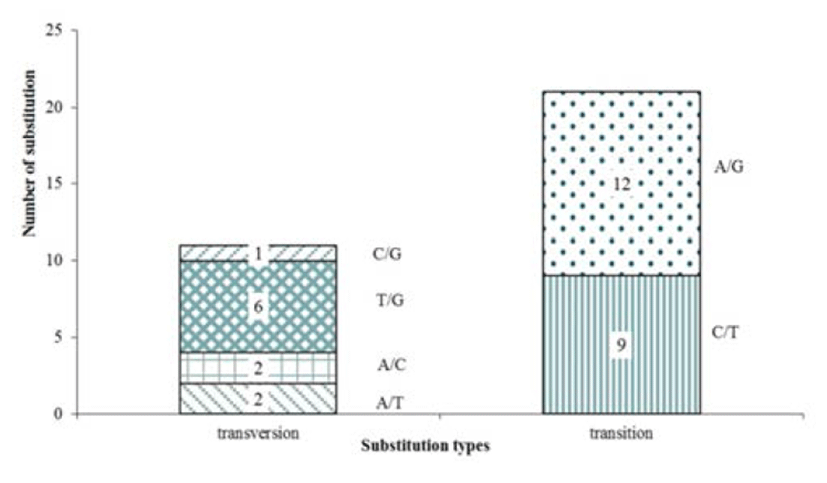
We were applied the designed primers on 16 sample (wild 8, cultured 8), and were performed PCR for amplifycation, with the result that 144 putative SNPs were selected for validation. Out of the 144 primer pairs, 25 primer did not amplify any product (17.4%) and 36 primer failed to sequencing (25.0%). Cause of failed sequencing is nonspecific bands (8, 5.6%), low PCR expression (15, 10.4%) and low alignment efficiency (13, 9.0%). But 83 markers (57.6%) were successfully analyzed in sequencing analysis. 51 out of 83 were confirmed monomorphic SNPs from the sample (wild 8, cultured 8), only 32 markers had detected polymorphic. 32 polymorphism SNP were observed in the variety genes region involved in immunity and protein synthesis (Table 4). The SNPs polymorphic are shown a total of 21 transitions (C/T and A/G), 11 transversions (A/T, A/C, T/G and C/G), and indel was not detected in polymorphic SNPs (Fig. 2). With C/T being the commonest (12) and C/G the least common (1) alteration observed. Transition/transversion ratio (ts/tv) was 1.910, SNP substitutions result of development marker shows transitions rate higher than transversions rate (Table 3). These results correspond with the EST resource SNP’s ones.
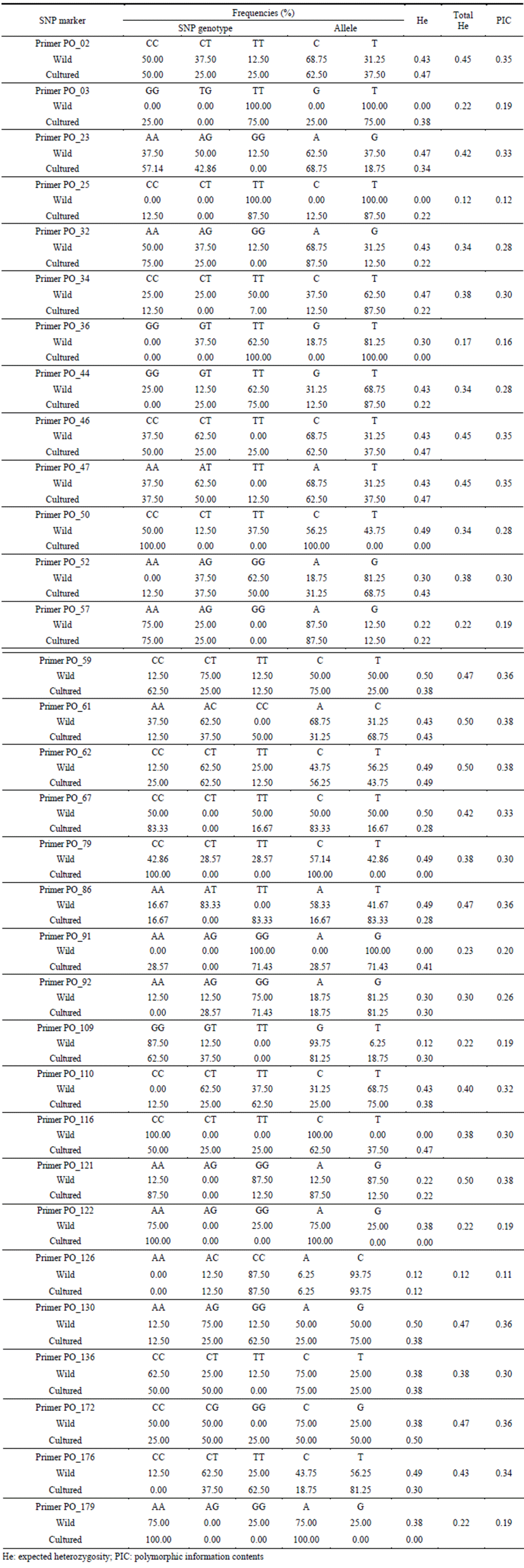
The calculation on frequency of genotyping and allele of 32 SNP markers, there were differences in allele frequency between wild and cultured olive flounder. In particular, Primer PO_03 (Allele: TT, 100%), Primer PO_25 (Allele: TT, 100%), Primer PO_91 (Allele: GG, 100%), Primer PO_116 (Allele: CC, 100%) marker had 100 percent one allele type in wild sample while cultured sample had various types. Meanwhile, Primer PO_36 (Allele: TT, 100%), Primer PO_50 (Allele: CC, 100%), Primer PO_79 (Allele: CC, 100%), Primer PO_122 (Allele: AA, 100%), Primer PO_179 (Allele: AA, 100%) had one allele type in cultured sample while had various alleles in wild sample. The analysis on heterozygosity (He) by sample showed 0.34 in wild sample and 0.29 in cultured sample, respecttively. The range of heterozygosity by marker was identified from 0.12 to 0.50. Primer PO_61, Primer PO_62, Primer PO_121 marker had the highest value of Heat 0.50 while Primer PO_126 marker had the lowest value of 0.12. The estimation of polymorphic information content (PIC) by marker ranged from 0.11 to 0.38 (Table 5).
DISCUSSIONS
Genomic regions of immune and protein synthesisrelated genes is directly related to life of organisms so that its importance is well known (He et al., 2011; He et al., 2012). In addition, single base substitution of specific genomic region can be a positive or negative a factor for survival, growth and adaptation to the environment. Because these usefulness, the SNP research has been actively proceeding in various fish species; but hasn’t yet been reported on olive flounder. In our experiments, discovery of SNP based on EST database in olive flounder and developed a valuable marker for applications to various genomic analyses. The SNPs polymorphism show that a total of 21 transitions (C/T and A/G), and 11 transversions (A/T, A/C, T/G and C/G) while transitions are higher than transversions. This result is similar with other previous research (Vera et al., 2011). These result suggest that transitions are more easily substituted than transversion, because purine-purine (transitions) substitution is easy and stable than purine-pyrimidine (transversions) substitution.
Moreover, 32 SNPs genomic region of identified polymorphism are related to immune response and protein synthesis, functional changes in the these proteins causes the phenotypic change. The single nucleotide substitution of the specific region might influences on immunity and survival of organisms. In particular, glutathione S-transferase, nephrosin, elastases factors, MHC II alpha beta antigens and apolipoprotein involved in innate immunity (Dijkstra et al., 2013; Tsai ea al., 2004; Vera et al., 2011). In addition, ferritin has an important role for iron storage and detoxification, the storage of iron is closely related to the immune response to bacterial infection protected. Finally, transferrin could be related to the inflammation and enhanced resistance to oxidative stress by parasite infections (Paramá et al., 2007). Also, SNP detected genes in olive flounder show a high similarity with immune-related genes in mammals (Vera et al., 2011). The innate immune response is an important and highly developed defence mechanism against pathogens in fish. Examples of innate immunity include anatomic barriers, mechanical removal of pathogens, bacterial antagonism, pattern-recognition receptors, antigen-nonspecific defense compounds, complement pathways, phagocytosis, and inflammation fish (Ellis et al., 2001). The research outcome can be used as basis data for a study to identify the relationship between the nucleotide substitution in the immune-related genes specific region and immune response.
The allele frequency analysis using newly developed SNP marker shows that a variety of alleles is present in wild olive flounder samples, but does not shows the diversity in a few cultured samples. It shows the necessity to use different markers for the wild and cultured sample for genome analysis. Primer PO_36, Primer PO_50, Primer PO_79, Primer PO_122, Primer PO_179 marker are useful to detect SNP for wild olive flounder samples, while Primer PO_03, Primer PO_25, Primer PO_91, Primer PO_116 are suitable for cultured samples. In addition, this specific marker is important to distinguish between wild and culture olive flounder sample.
The diversity analysis using developed SNP marker in our research shows the heterozygosity value is higher in the wild sample of 0.34 than the cultured sample of 0.29, meaning higher diversity of the wild than the cultured. The cultured sample has got smaller diversity due to continuous inbreeding in the group without any inflow of new species (new mother group), resulting in lower diversity than the wild (Xing et al., 2014). Result of our heterozygosity is a correlation with diversity study outcome of olive flounder using microsatellite (Jung et al., 2009). It supports the appropriateness and usefulness of newly developed SNP marker to apply not only to SNP detection but also to other diversity analysis.
In conclusion, we was identified the substitution using the new marker from sample, it supports that development marker on this study is suitable for SNP detection in olive flounder. In addition, the marker is an appropriate tool for diversity analysis. The outcome of this study will be used in the selection of breeding with economic traits and will facilitate the molecular breeding programs of olive flounder.