ARTICLE
Genetic Distances and Variations of Three Clupeid Species Determined by PCR Technique
Sang-Hoon Choi, Jong-Man Yoon†
†Corresponding Author : Jong-Man Yoon, Dept. of Aquatic Life Medicine, College of Ocean Science and Technology, Kunsan National University, Gunsan 573-701, Korea. Tel. : +82-63-469-1887, Fax : +82-63-469-1887, E-mail :
jmyoon@kunsan.ac.kr
Copyright © 2014 © Copyright an Official Journal of the Korean Society of Developmental Biology. All Rights Reserve. This is an Open-Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received: Dec 1, 2014 ; Revised: Dec 5, 2014 ; Accepted: Dec 8, 2014
ABSTRACT
In this study, seven oligonucleotides primers were shown to generate the shared loci, specific loci, unique shared loci to each species and shared loci by the three species which could be obviously calculated. Euclidean genetic distances within- and between-species were also calculated by complete linkage method with the sustenance of the hierarchical dendrogram program Systat version 13. The genomic DNA isolated from herring (Clupea pallasii), Korean anchovy (Coilia nasus) and large-eyed herring (Harengula zunashi), respectively, in the Yellow Sea, were amplified several times by PCR reaction. The hierarchical dendrogram shows three chief branches: cluster 1 (PALLASII 01, 02, 03, 04, 06 and 07), cluster 2 (NASUS 08, 09, 10, 11, 12, 13 and 14), and cluster 3 (ZUNASHI 15, 16, 17, 18, 19, 20, 21 and PALLASII 05). In three clupeid species, the shortest genetic distance displaying significant molecular difference was between individual PALLASII no. 03 and PALLASII no. 02 (0.018). Individual no. 06 of PALLASII was most distantly related to NASUS no. 11 (genetic distance = 0.318). Individuals from herring (C. pallasii) species (0.920) exhibited higher bandsharing values than did individuals from Korean anchovy (C. nasus) species (0.872) (P<0.05). As a result, this PCR analysis generated on the genetic data displayed that the herring (C. pallasii) species was widely separated from Korean anchovy (C. nasus) species. Reversely, individuals of Korean anchovy (C. nasus) species were a little closely related to those of large-eyed herring (H. zunashi) species.
Keywords: Clupeid; Clupea nasus; Clupea pallasii; Euclidean genetic distance; Harengula zunashi
INTRODUCTION
The authors have investigated three species of Clupeidae such as herring (Clupea pallasii), Korean anchovy (Coilia nasus) and large-eyed herring (Harengula zunashi), belonging to the order Clupeiformes. The fish is also indigenous to some parts of the southern regions of the Yellow Sea. Fishes are the most popular marine products in Korea because of their taste and nutritional value, and Koreans consume them in large quantities. Especially, Korean anchovy and large-eyed herring are widely distributed in the entirety of brackish-water habitats and seawater areas of the Yellow Sea in the Korean Peninsula, as well as in several areas in China. However, in spite of their economic and scientific consequences, a little information currently exist regarding the taxonomical (Kim et al., 2001), physical (An et al., 2013) and nutritional (Heu et al., 2012) levels only of clupeid species in Korea. During the last two decades, environmental contamination and various environmental disruptions by industries and city sewage, have threatened the coastal fisheries, and then reduction of individual number of this fish is an increasing trend in the 2000s. This study attempt is to elucidate the genetic distances and variations within and between herring, Korean anchovy and large-eyed herring species from the Yellow Sea of the Korean Peninsula.
MATERIALS AND METHODS
Muscle tissues were obtained separately from individuals of three clupeid species such as herring (C. pallasii), Korean anchovy (C. nasus) and large-eyed herring (H. zunashi) from the Yellow Sea of the Korean Peninsula, respectively. Three species of fish muscle was collected in sterile tubes, placed on ice immediately, and stored under refrigeration until needed. Genomic DNA was extracted and purified under the conditions described previously (Yoon & Kim, 2004; Song & Yoon, 2013). The DNA pellets were incubation-dried for 2 hrs, held at –40°C until analysis, and then dissolved in the TE buffer (10 mM Tris- HCl, pH 8.0; 1 mM EDTA). The concentrations of the extracted genomic DNA samples were estimated based on the absorbance at 260 nm by a spectrophotometer (Beckman Coulter, Buckinghamshire, UK). PCR analysis was performed on DNA samples extracted from a total of 21 individuals using seven decamer primers. Seven oligonucleotides primers (BION-07, BION-10, BION-11, BION-18, BION-25, BION- 27 and BION-40; Bioneer Co. Daejeon, Korea) were displayed to generate the shared loci, specific loci, unique shared loci to each species and shared loci by the three species which could be obviously scored. The degree of variability was calculated by use of the Dice coefficient (F), which is given by the formula: F = 2 nab / (na+nb), where nab is the number of bands shared between the samples a and b, na is the total number of bands for sample a and nb is the total number of bands for sample b (Jeffreys & Morton, 1987; Yoke-Kqueen & Radu, 2006). Euclidean genetic distances within- and between-species were also calculated by complete linkage method with the support of the hierarchical dendrogram program Systat version 13 (SPSS Inc., Chicago, IL, USA).
RESULTS AND DISCUSSION
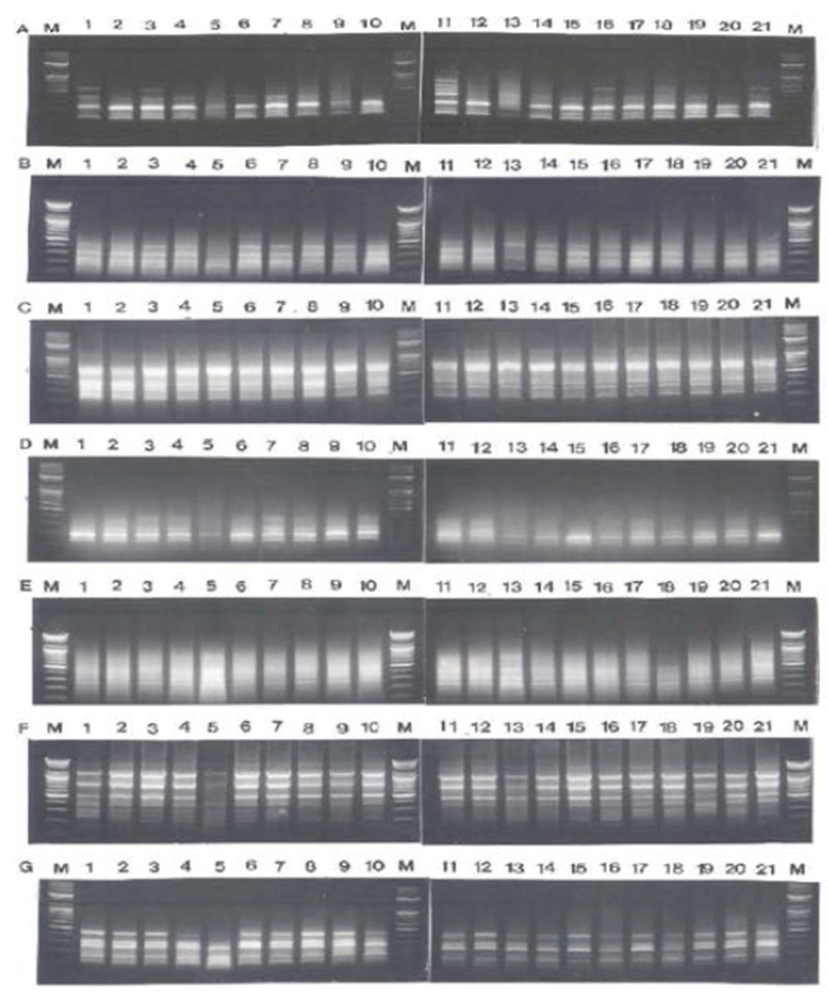
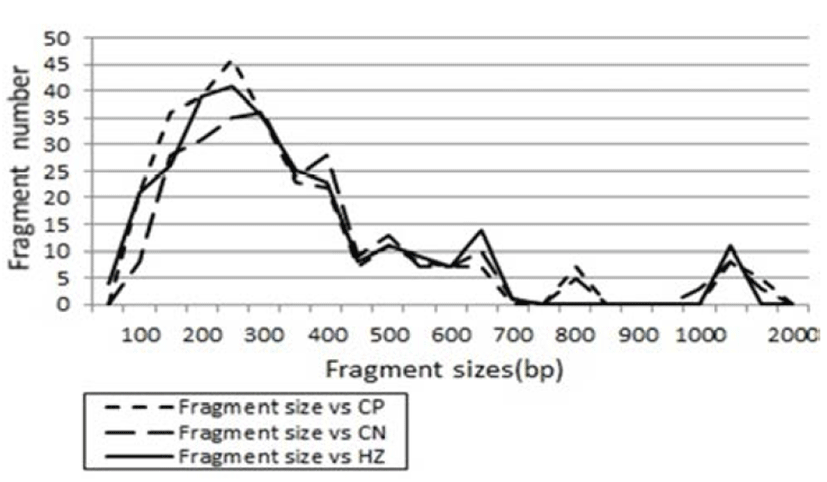
The genomic DNA isolated from herring (C. pallasii), Korean anchovy (C. nasus) and large-eyed herring (H. zunashi) in the Yellow Sea, respectively, were amplified several times by PCR reaction, as shown in Fig. 1. Similarity matrix including bandsharing values (BS) and genetic differences was calculated using Nei and Li's index of the similarity of clupeid individuals from the Yellow Sea of the Korean Peninsula, respectively. The band-sharing value between individuals no. 02 and no. 03 was 0.976 within the herring species (C. pallasii), which was the highest value identified among the three species (unpublished data). In the present study, averagely, an oligonucleotides primer generated 40.3 amplified products per primer in the herring species, 37.9 in Korean anchovy species and 39.3 in large-eyed herring species, as demon-strated in Table 1. Here, the seven oligonucleotides primers BION-07, BION-10, BION-11, BION-18, BION-25, BION-27 and BION-40 were used to generate the unique shared loci to each species and shared loci by the three species, as illustrated in Table 2. 259 unique shared loci to each species, with an average of 37 per primer, were observed in the herring species, and 217 loci, with an average of 31 per primer, were observed in the Korean anchovy species. As regards average bandsharing value (BS) results, indi-viduals from herring (C. pallasii) species (0.920) exhibited higher bandsharing values than did individuals from Korean anchovy (C. nasus) species (0.872) (P<0.05), as illustrated in Table 3. DNA fragments attained by seven decamer primers ranged in size from 50 bp to 2,200 bp in the three clupeid species, as displayed in Fig. 2. The higher fragment sizes (>1,000 bp) are much more observed in the herring species. Many researchers studied the sizes of DNA fragments in the PCR profiles of barramundi (Lates calcarifer) (Partis & Wells, 1996), four species of the Mullidae family (Mamuris et al., 1999), the eastern Pacific yellowfin tuna (Thunnus albacares) (Diaz-Jaimes & Uribe-Alcocer, 2003), five species of Korean catfish (Silurus asotus) (Yoon & Kim, 2004), venus clam (Gomphina aequilatera) (Kim et al., 2006) and the turtle leg (Pollicipes mitella) (Song & Yoon, 2013).
Table 3.
Multiple comparisons of average bandsharing values among herring (C. pallasii), Korean anchovy (C. nasus) and large-eyed herring (H. zunashi) species, respectively, were generated according to the bandsharing values
|
Species |
CP |
CN |
HZ |
|
CP |
0.920±0.081a
|
0.763±0.048cd
|
0.770±0.055cd
|
|
CN |
- |
0.872±0.091b
|
0.704±0.054e
|
|
HZ |
- |
- |
0.890±0.090b
|
Download Excel Table
Table 2.
The number of unique loci to each species and number of shared loci by the three species generated by PCR analysis using 7 oligonucleotides primers in herring (C. pallasii), Korean anchovy (C. nasus) and large-eyed herring (H. zunashi) species, respectively
|
Item |
No. of unique loci to each species |
No. of shared loci by the three species |
|
Primer \ species |
CP |
CN |
HZ |
Three species (7 individuals per species) |
|
BION-07 |
49 |
49 |
56 |
126 |
|
BION-10 |
42 |
28 |
42 |
63 |
|
BION-11 |
42 |
42 |
35 |
63 |
|
BION-18 |
42 |
21 |
49 |
63 |
|
BION-25 |
35 |
35 |
49 |
84 |
|
BION-27 |
28 |
28 |
14 |
21 |
|
BION-40 |
21 |
14 |
14 |
42 |
|
Total no. |
259 |
217 |
259 |
462 |
|
Average no. per primer |
37 |
31 |
37 |
66 |
Download Excel Table
Table 1.
The number of average loci per lane and specific loci by PCR inquiry using 7 oligonucleotides primers in herring (C. pallasii), Korean anchovy (C. nasus) and large-eyed herring (H. zunashi) species
|
Item |
No. of average loci per lane |
No. of specific loci |
|
Primer |
CP |
CN |
HZ |
CP |
CN |
HZ |
|
BION-07 |
7.14(50) |
8.14(5.7) |
8.57(60) |
1 |
8 |
4 |
|
BION-10 |
5.57(39) |
4.71(33) |
5.29(37) |
18 |
26 |
16 |
|
BION-11 |
3.86(27) |
3.14(22) |
3.14(22) |
6 |
1 |
8 |
|
BION-18 |
7.57(53) |
6()42 |
7(49) |
11 |
21 |
0 |
|
BION-25 |
6.71(47) |
7.43(52) |
7(49) |
12 |
10 |
0 |
|
BION-27 |
4.89(34) |
4.57(32) |
5.57(39) |
6 |
4 |
4 |
|
BION-40 |
4.57(32) |
3.86(27) |
2.71(19) |
11 |
11 |
5 |
|
Total no. |
40.3(28.2) |
37.9(265) |
39.3(275) |
65 |
81 |
37 |
|
Average no. per primer |
40.3 |
37.9 |
39.3 |
9.3 |
11.6 |
5.3 |
Download Excel Table
Fig. 1.
PCR-based electrophoretic profiles of C. pallasii (lane 1-7), C. nasus (lane 8-14) and H. zunashi (lane 15-21), respectively, from the Yellow Sea of the Korean Peninsula, amplified by oligonucleotides primers BION-07 (A), BION-10 (B), BION-11 (C), BION-18 (D), BION-25 (E), BION- 27 (F) and BION-40 (G). Each lane shows differrent individual DNA samples used. Amplified products were electrophoresed on a 1.4% agarose gel and detected by staining with ethidium bromide. 100 bp ladder markers were used as a DNA molecular size marker (M).
Download Original Figure
Fig. 2.
Distribution of fragment sizes of herring (C. pallasii), Korean anchovy (C. nasus) and largeeyed herring (H. zunashi) species, respectively. Solid lines: HZ species. Dotted line: CP species. Thick dotted lines: CN species. The fragment numbers in each size interval have been computed from the pooled fragments obtained with all the primers. The higher fragment sizes (>1,000 bp) are much more observed in the HZ species.
Download Original Figure
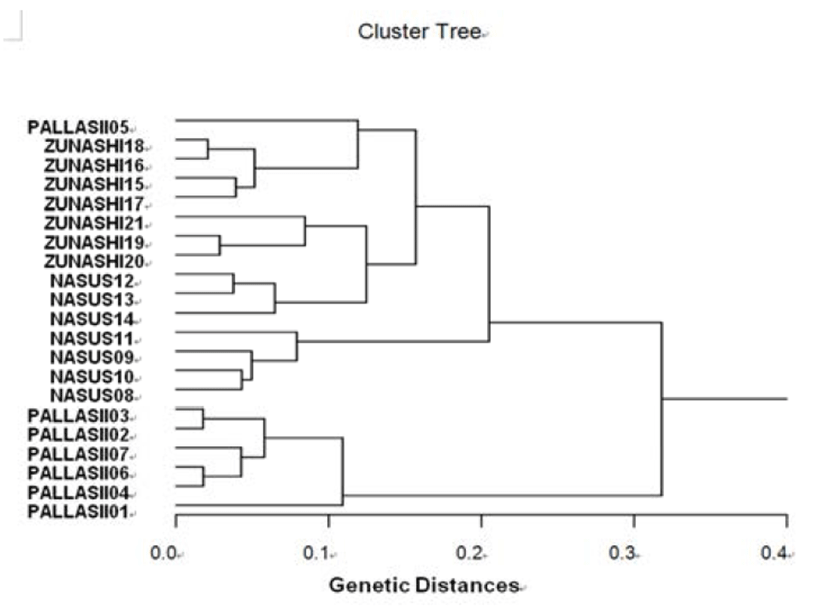
The hierarchical dendrogram indicates three main branches: cluster 1 (PALLASII 01, 02, 03, 04, 06 and 07), cluster 2 (NASUS 08, 09, 10, 11, 12, 13 and 14), and cluster 3 (ZUNASHI 15, 16, 17, 18, 19, 20, 21 and PALLASII 05) (Fig. 3). The genetic distance among the three clupeid species ranged from 0.018 to 0.318. In three clupeid species, the shortest genetic distance displaying significant molecular difference was between individual PALLASII no. 03 and PALLASII no. 02 (0.018). In the long run, individual no. 06 of the PALLASII was most distantly related to NASUS no. 11 (genetic distance = 0.318). DNA fragments identified in this study may be useful as a DNA marker. The values of the pairwise comparisons of unbiased genetic distance between the populations of the Indian major carp (Catla catla) from the combined data for the four primers, ranged from 0.025 to 0.052 (Islam et al., 2005). They reported that the Padma and the Jamuna populations were separated from each other with the lowest genetic distance (D = 0.025). The genetic distance between the Indian Ocean lobster and the Korean slipper lobster species ranged between 0.040 and 0.612 (Park et al., 2005). They suggested that this genetic technique could be used to discriminate between different river populations of major carp.
Fig. 3.
Hierarchical dendrogram of genetic distances, accomplished from three clupeid species. The related construction among different individuals from herring (PALLASII), Korean anchovy (NASUS) and large-eyed herring (ZUNASHI) species, respectively, was created according to the bandsharing values and similarity matrix.
Download Original Figure
Consequently, PCR analysis generated on the genetic data displayed that the herring (C. pallasii) species was widely separated from Korean anchovy (C. nasus) species. Reversely, individuals of Korean anchovy species were properly closely related to those of large-eyed herring species, as shown in the hierarchical dendrogram of genetic distances. As stated above, the potential of genetic analysis to identify diagnostic markers for the identification of three clupeid species has been demonstrated (McCormack et al., 2000). Generally speaking, this PCR analysis method has been applied to identify specific markers particular to line, species and geographical population, as well as genetic diversity/polymorphism in diverse species of organisms (McCormack et al., 2000; Kim et al., 2006; Yoon & Kim, 2004).
ACKNOWLEDGEMENTS
Particular thanks go to Dae-Hyun Kim, Jun-Hyub Jeon and my laboratory colleagues for their assistance in sample collection and their backing with the procedure of PCR technique and statistical analysis. The authors thank also to referees who assisted with thorough and profound correction.
REFERENCES
An HC, Kim SH, Lim JH, Bae JH. Catching efficiency of the biodegradable gill net for Pacific herring (
Clupea pallasii). Bull Korean Soc Fish Technol. 2013; 49:341-351.

Diaz-Jaimes P, Uribe-Alcocer M. Allozyme and RAPD variation in the eastern Pacific yellowfin tuna (
Thunnus albacares). Fish Bull. 2003; 101:769-777.

Heu MS, Park KH, Shin JH, Lee JS, Yeum DM, Lee DH, Kim HJ, Kim JS. Sanitary and nutritional characterization of commercial Kwamegi from pacific herring
Clupea pallasii. Korean J Fish Aquat Sci. 2012; 45:1-10.

Islam MS, Ahmed ASI, Azam MS, Alam MS. Genetic analysis of three river populations of
Catla catla (HAMILTON) using randomly amplified polymorphic DNAs markers. Asian-Aust J Anim Sci. 2005; 18:453-457.

Jeffreys AJ, Morton DB. DNA fingerprints of dogs and cats. Anim Genet. 1987; 18:1-15.

Kim JK, Kang CB, Han KH, Kim YU. A new record of the herring,
Sardinella lemuru (Pisces Clupeidae) from Korea. Korean J Ichthyol. 2001; 13:190-194.

Kim Jr, Jung CH, Kim YH, Yoon JM. Genetic variations in geographic venus clam (
Gomphina aequilatera, Sowerby) populations from Samcheok and Wonsan. Dev Reprod. 2006; 10:227-238.

Mamuris Z, Stamatis C, Bani M, Triantaphyllidis C. Taxonomic relationships between four species of the Mullidae family revealed by three genetic methods allozymes, random amplified polymorphic DNA and mitochondrial DNA. J Fish Biol. 1999; 55:572-587.

McCormack GC, Powell R, Keegan B. Comparative analysis of two populations of the brittle star
Amphiura filiformis (Echinodermata Ophiuroidae) with different life history strategies using RAPD markers. Mar Biotechnol. 2000; 2:100-106.

Park SY, Park JS and Yoon JM. Genetic differences and variations in slipper lobster (
Ibacus ciliatus) and deep sea lobster (
Puerulus sewelli) determined by RAPD analysis. Korean J Genet. 2005; 25:307-317.

Partis L, Wells RJ. Identification of fish species using random amplified polymorphic DNA (RAPD). Mol Cell Probes. 1996; 10:435-441.

Song YJ, Yoon JM. Genetic differences of three
Pollicipes mitella populations identified by PCR analysis. ev Reprod. 2013; 17:199-205.

Yoke-Kqueen C, Radu S. Random amplified polymorphic DNA analysis of genetically modified organisms. J Biotechnol. 2006; 127:161-166.

Yoon JM, Kim JY. Genetic differences within and between populations of Korean catfish (
S.asotus) and bullhead (
P. fulvidraco) analysed by RAPD-PCR. Asian- Aust J Anim Sci. 2004; 17:1053-1061.