INTRODUCTION
The ability to generate induced pluripotent stem cells (iPSCs) using a set of four defined transcription factors (Oct4, Sox2, Klf4, and c-Myc (OSKM)) provides great promise for generating patient-specific cells that are suitable for cell therapy, disease modeling, and drug screening (Chi et al., 2015; Tidball and Parent, 2016; Yamashita and Abe, 2016). Despite this great promise, reprogramming efficiencies in initial studies with the OSKM transcription factors were low (0.01–0.02%) and are probably not sufficient for clinical applications (Ebrahimi, 2015).
Therefore, diverse strategies have been developed to improve the reprogramming efficiency, including the choice of various reprogramming factors, microRNAs, and small molecules and alteration of the culture conditions (Li and Rana, 2012; Luni et al., 2016; Onder et al., 2012; Unternaehrer et al., 2014). These approaches target the main chromatin-remodeling pathways as well as cellular signaling pathways.
Meanwhile, recent studies revealed that iPSCs generated by either integrative or non-integrative methods have genetic abnormalities (Gore et al., 2011; Hussein et al., 2011; Laurent et al., 2011). During the process of reprogramming, the levels of phosphorylated histone H2AX (γH2AX), one of the earliest indictors of DNA double-strand breaks (DSBs), increase greatly, indicating that reprogramming is accompanied by severe genomic DNA damage (Gonzalez et al., 2013). Indeed, reprogramming factors induce apoptotic signals through the accumulation of oxidative stress and DNA damage (Banito et al., 2009>; Kawamura et al., 2009). A study of the DNA repair mechanism during reprogramming of somatic cells into a pluripotent statefurther confirmed that the primary source of genetic instability is DNA DSBs that arise during the process of reprogramming (Gonzalez et al., 2013). DNA DSBs are repaired by homologous recombination (HR) or non-homologousend-joining (NHEJ). Defects in the major components of HR or NHEJ prevent the generation of iPSCs; therefore, an intact HR or NHEJ pathway is considered to be essential for efficient reprogramming (Felgentreff et al., 2014; Gonzalez et al., 2013).
Rad51 is a key component of HR and it forms Rad51 recombinase filaments of broken single-stranded DNA to promote HR (Sugawara et al., 2003; White and Haber, 1990). In addition, Rad51 regulates cell cycle progression in mouse embryonic stem cells by preserving G2/M transition (Yoon et al., 2014). Rad51 also facilitates replication fork progression by inhibiting replication fork collapse to ensure cell proliferation in vertebrates (Sonoda et al., 1998; Tsuzuki et al., 1996). Taken together, Rad51 is considered to play a critical role inprotecting genome integrity as well as resistance against apoptosis triggered by DNA damage both in stem cells and somatic cells. When expression of Rad51 is suppressed, the reprogramming efficiency is significantly decreased, indicating that Rad51 plays a critical role in reprogramming into a pluripotent state (Gonzalez et al., 2013).
In this study, we demonstrate that Rad51 enhances the reprogramming efficiency during the reprogramming of mouse embryonic fibroblasts (MEFs) into iPSCs. We further suggest that Rad51 facilitates the DNA damage response and mesenchymal-to-epithelial transition.
MATERIALS AND METHODS
MEFs expressing green fluorescent protein (GFP) from the Oct4 locus (OG-MEFs, passage 2) were purchased (Stemgent, MA, USA). To induced pluripotency, 4 × 105 cells were seeded on a 15 cm cell culture dish coated with 0.2% gelatin. The cells were transduced with pMx-based retroviral reprogramming factors. Twenty hours after transduction, the cells were transferred to J1 mouse embryonic stemcell growth media and maintained as described previously (Jirmanova et al., 2002).
Total RNA was extracted using TRIzol (Invitrogen, CA, USA), and 2–5 µg of total RNA was reverse-transcribed using the SuperScriptII™ First-Strand Synthesis System (Invitrogen) according to the manufacturer’s instructions. Real-time RT-PCR was performed using cDNA with the QuantiTectSYBR Green PCR Kit (Qiagen, CA, USA). Reactions were performed in triplicate using an Exicycler™ 96 real-time quantitative thermal block (Bioneer, Daejon, Korea). For quantification, the expression levels of the target genes were normalized against those of the glyceraldehyde 3-phosphate dehydrogenase gene.
Whole-cell extracts were prepared, and 20−50 μg of protein was resolved by SDS-PAGE and blotted using antibodies against Rad51 (sc-8349; Santa Cruz, CA, USA), γH2AX (ab22551; Abcam, CA, USA) and β-actin (sc-47778, Santa Cruz). Immunoreactivity was detected by enhanced chemiluminescence (Amersham, Buckinghamshire, England).
Graphical data are presented as means ± standard deviation. Each experiment was performed at least three times and subjected to statistical analysis. Statistical significance between two groups was determined using the Student’s t-test. A p value less than 0.05 was considered significant. Statistical analysis was performed using the SAS statistical package v.9.13 (SAS Inc, NC, USA).
RESULTS AND DISCUSSION
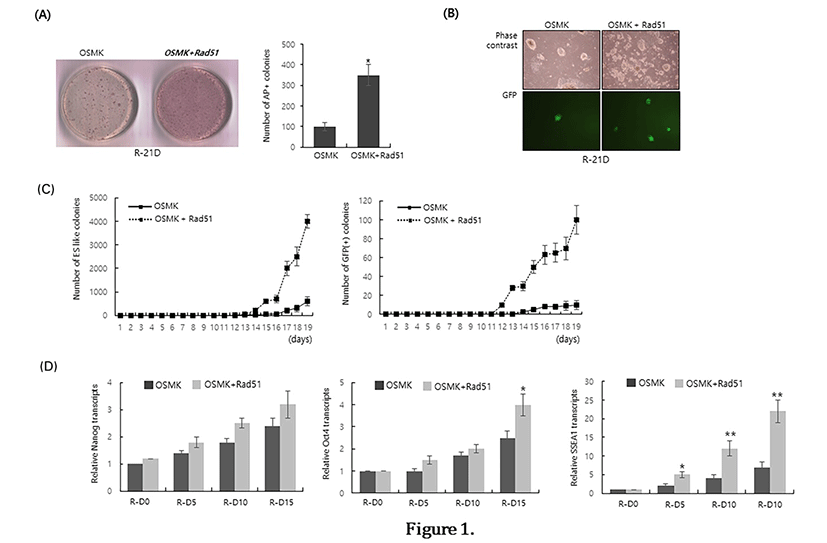
To examine the effect of Rad51 expression on the reprogramming efficiency of MEFs, we co-transduced a Rad51-expressing retrovirus with four reprogramming factors (OSKM) and compared the number of alkaline phosphatase-positive (AP(+)) colonies at 21 days after reprogramming initiation. Co-expression of Rad51 increased the number of AP(+) colonies by approximately 3.5-fold compared with OSKM transduction alone (Fig. 1A). To further confirm that Rad51 enhances the reprogramming of MEFs, we reprogrammed reporter MEFs expressing GFP under the control of the Oct4 promoter (OG-MEFs). Consistent with the results of alkaline phosphatasestaining, co-expression of Rad51 and OSKM enhanced the number of colonies expressing GFP, which is a marker of activation of the Oct4 promoter by reprogramming (Fig. 1B). We next counted the numbers of ES-like colonies and GFP-expressing colonies during the reprogramming of OG-MEFs to examine whether Rad51 expression facilitates the reprogramming process. GFP-positive colonies as well as ES-like colonies appeared at least 3–4 days earlier when Rad51 was co-expressed with OSKM (Fig. 1C). Even though Nanog expression was not significantly increased in the presence of Rad51 until 15 days after the initiation of reprogramming, the expression of Oct4 and SSEA1 was higher when Rad51 was co-expressed with reprogramming 4 factors (Fig. 1D). These results suggest that co-expression of Rad51and OSKM can facilitate the reprogramming process and enhance the reprogramming efficiency of MEFs.
Reprogramming is a process of mesenchymal-to-epi-thelial transition, which isaccompanied by upregulation of epithelial factors and down regulation of mesenchymal factors Samavarchi-Tehrani et al., 2010). Therefore, we investigated the effect of Rad51 on the expression of epithelial and mesenchymal markers during the process of reprogramming. As expected, expression of Snail, a mesenchymal marker, was decreased, whereas expression of epithelial markers such as E-cadherin and epithelial cell adhesion molecule (Ep-CAM) was increased at the early stage of reprogramming (Fig. 2A). Compared with OSKM transduction alone, co-expression of Rad51 significantly increased E-cadherin and Ep-CAM expression from 6 days after reprogramming initiation, even though Rad51 expression did not have any effect on the expression of Snail (Fig. 2A).
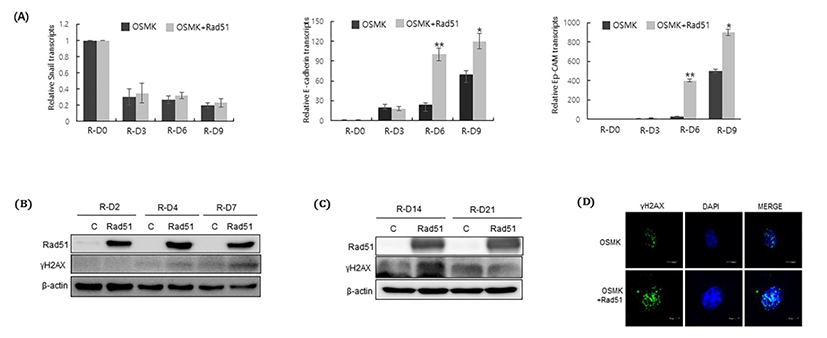
Rad51 is a DNA repair protein that mediatesHR of DNA DSBs; therefore, we next examined whether co-expression of Rad51 modulates the DNA damage repair system during reprogramming. Phosphorylation of histone H2AX is essential for promoting DNA DSB repair by initiating the repair system (Kinner et al., 2008). Therefore, we compared the level of γH2AX in the presence and absence of Rad51 during the process of reprogramming. The level ofγH2AX was increased in the presence of Rad51 during the early stage of reprogramming (Fig. 2B). To further confirm that γH2AX accumulates, we examined the level of γH2AX at 14 and 21days after reprogramming initiation. The level of γH2AX was highly increased after 14 days of reprogramming in the presence of Rad51 (Fig. 2C). Immunocytochemical staining further showed that the level of γH2AX was high after 14 days of reprogramming (Fig. 2D). Taken together, we concluded that the ability of Rad51 to enhance the reprogramming efficiency is associated with promotion of the DNA repair mechanism during the reprogramming process.
In this study, we provide compelling evidence that Rad51 plays an important role inincreasing the reprogramming efficiency of MEFs. The mutation rate of pluripotent stem cells are relatively low compared with somatic cells at least in part by the active operation of HR pathway to repair DNA damage (Chlon et al., 2016). The molecular mechanisms by which Rad51 activity enhances the reprogramming efficiency remain obscure, although we speculate that the primary mechanism is activation of the DNA damage response. Indeed, γH2AX began to accumulate early during reprogramming when Rad51was co-expressed with OSKM. This result indirectly indicates that expression of Rad51 facilitates activation of the HR repair system.
Another possible mechanism by which Rad51 activity enhances the reprogramming efficiency is through regulation of the cell cycle. Cell cycle progression is closely associated with the efficiency of reprogramming, and the reprogramming factor c-Myc plays a central role inestablishing the pluripotent cell-specific cell cycle structure (Singh and Dalton, 2009).
In addition to its activity in HR, Rad51preserves cell cycle progression by preventing DNA replication fork collapse and mediates replication restart by functioning at stalled replication forks (Somyajit et al., 2015). Given that the reprogramming process isaccompanied by severe replication stress, it is also reasonable to hypothesize that Rad51 might be able to facilitate cell cycle progression by reducing replication stress during the reprogramming process.
The genomic instability of iPSCs has raised concerns about their clinical use; therefore, it is essential to determine whether co-expression of Rad51 and OSKM increases the genomic stability of established iPSCs, in addition to enhancing the reprogramming efficiency.
