INTRODUCTION
In the present study, three ascidian species such as Styela clava (SC), Halocynthia roretzi (HR) and Styela plicata (SP), belonging to the class Ascidiacea, are the most popular marine products in Korea because of their scent, flavor and nourishing value, and Koreans consume them plentifully. These ascidians, economically important marine species broadly distributed on the coast of southern sea of the Korean peninsula and the several sea areas in the natural ecosystem. However, notwithstanding their economic and scientific significances, a little information currently exists concerning the stability of producibility (Huh & Choi, 2008), the physiology (Shin et al., 2007) and the development of aquacultural technique (Lee, 2011) points only of ascidian species in Korea. Also, during the last two decades, parasite outbreak and various environmental disruptions by industries, city sewage and global warming, have threatened the coastal fisheries, and then reduction of individual number of this invertebrate is an increasing trend in the 2000s. Among various PCR-based molecular techniques, RAPDs are the most frequently used molecular markers for taxonomic and systematic analyses of organisms (Adams, 2000; Bartish et al., 2000). Particularly, the polymorphic and/or specific markers specific to the breed, the species, the genus or the geographical population have been applied for the identification of individuals and population, hybrid parentage and for the screening of DNA (Guo et al., 2001). Generally, RAPD-PCR is one of fast and simple research methods to identify genetic difference and the polymorphism in various organisms that does not require the prior knowledge of the genomic DNA (Iyengar et al., 2000; Nebauer et al., 2000; Nicolosi et al., 2000). In this study, to explicate the genetic distances among ascidian species, the author achieved a clustering analysis of three ascidian species collected from the southern sea.
MATERIALS AND METHODS
Muscle tissues were obtained separately from individuals of three ascidian species such as Styela clava (SC), Halocynthia roretzi (HR) and Styela plicata (SP) from the southern sea of the Korean Peninsula, respectively. The muscle was collected in sterile tubes, immediately placed on dry ice, and stored at under freezing until the genomic DNA extraction. Genomic DNA was extracted and purified under the conditions designated previously (Yoon & Kim, 2004; Song & Yoon, 2013). The DNA pellets were incubationdried for 2 hrs, held at –40°C until analysis, and then dissolved in the TE buffer (10 mM Tris-HCl, pH 8.0; 1 mM EDTA). The concentrations of the extracted genomic DNA samples were estimated based on the absorbance at 260 nm by a spectrophotometer (Beckman Coulter, Buckinghamshire, UK). PCR analysis was accomplished on DNA samples extracted from a total of 21 individuals using seven oligonucleotides primers. Seven oligonucleotides primers BION-11 (5'-GTGATCGCAG-3'), BION-13 (5'-GTTTCG CTCC-3'), BION-17 (5'-TGCTCTGCCC-3'), BION-21 (5'-AATCGGGCTG-3'), BION-23 (5'-CAGGCGGCGT-3'), BION- 25 (5'-CTGGCGGCTG-3') BION-30 (5'-GCCACCTCCT-3') were demonstrated to produce the total loci, average loci per lane, specific loci, unique loci to each species and shared loci by the three species which could be clearly counted. The degree of variability was estimated by use of the Dice coefficient (F), which is given by the formula: F = 2 nab / (na+nb), where nab is the number of bands shared between the samples a and b, na is the total number of bands for sample a and nb is the total number of bands for sample b (Jeffreys & Morton, 1987; Yoke-Kqueen & Radu, 2006). Euclidean genetic distances within- and between-species were also calculated by complete linkage method with the support of the hierarchical dendrogram program Systat version 13 (SPSS Inc., Chicago, IL, USA).
RESULTS AND DISCUSSION
The genomic DNA isolated from three ascidians such as Styela clava (SC), Halocynthia roretzi (HR) and Styela plicata (SP) in the southern sea, respectively, were amplified several times by PCR reaction (unpublished data). Similarity matrix including bandsharing values (BS) was calculated using Nei and Li's index of the similarity of ascidian individuals from the southern sea of the Korean Peninsula, respectively, as demonstrated in Table 1. Based on the average bandsharing values of all samples, the similarity matrix ranged from 0.519 to 0.774 in the SC species, from 0.261 to 0.683 in the HR species and from 0.346 to 0.730 in the SP species, as illustrated in Table 1. The bandsharing value between individuals no. 01 and no. 02 was 0.774, which was the highest value identified within the SC species. Seven oligonucleotides primers were shown to generate the shared loci, specific loci, which could be obviously scored, as demonstrated in Table 2. In the present study, 7 oligonucleotides primers produced 401 total loci in the SC, 390 in the HR and 434 in the SP species, respectively. Also, these oligonucleotides primers produced 275 specific loci in the SC, 341 in the HR and 364 in the SP species, respectively. The specific loci generated by oligonucleotides
Bandsharing values of three ascidian species are above the diagonal primers exhibited inter-species-specific characteristics, thus revealing DNA polymorphisms. Here, the seven oligo-nucleotides primers were used to generate the unique loci to each species and shared loci by the three species, as illustrated in Table 3. 126 unique shared loci to each species, with an average of 18 per primer, were observed in the SC species, 49 loci, with an average of 7 per primer, were observed in the HR species, and 70 loci, with an average of 10 per primer, and were observed in the SP species.
The oligonucleotides primer BION-17 generated 28 unique loci to each species in the SC species. The oligonucleotides primer BION-23 generated 28 unique loci to each species in the SP species. Specifically, the oligo-nucleotides primer BION-13 generated 7 unique loci to each species, which were identifying each species in the HR species. Especially, the oligonucleotides primer BION-25 generated 7 unique loci to each species, which were identifying each species in the SP species. Interestingly, BION-17 distinguished 21 shared loci by the three ascidian species, major and/or minor fragments of sizes, which were identical in almost all of the samples. Also, the primer BION-23 detected 21 shared loci by the three species, various fragments of sizes, which were identical in almost all of the samples. As regards average bandsharing value (BS) results, individuals from SC species (0.661±0.081) exhibited higher bandsharing values than did individuals from HR species (0.555±0.074) (P<0.05), as illustrated in Table 4. The dendrogram resulted from trustworthy seven oligonucleotides primers, indicating three genetic groups composed of cluster I (CLAVA 01, 02, 03, 04, 05, 06 and 07), cluster II (RORETZI 08, 09, 10, 11, 12, 13 and 14) and cluster III (PLICATA 15, 16, 17, 18, 19, 20 and 21), as shown in Fig. 1. The genetic distance among the three ascidian species ranged from 0.071 to 0.599. In three ascidian species, the longest genetic distance (0.599) exhibiting significant molecular difference was also between individual no. 02 within SC species and individual no. 14 within HR species. The shortest genetic distance (0.071) exhibiting significant molecular difference was also between individual no. 20 and no. 21 within the SP species.
| Species | SC | HR | SP |
|---|---|---|---|
| SC | 0.661±0.081 d | 0.459±0.072 a | 0.508±0.073 b |
| HR | - | 0.555±0.074 c | 0.495±0.044 b |
| SC | - | - | 0.587±0.069 c |
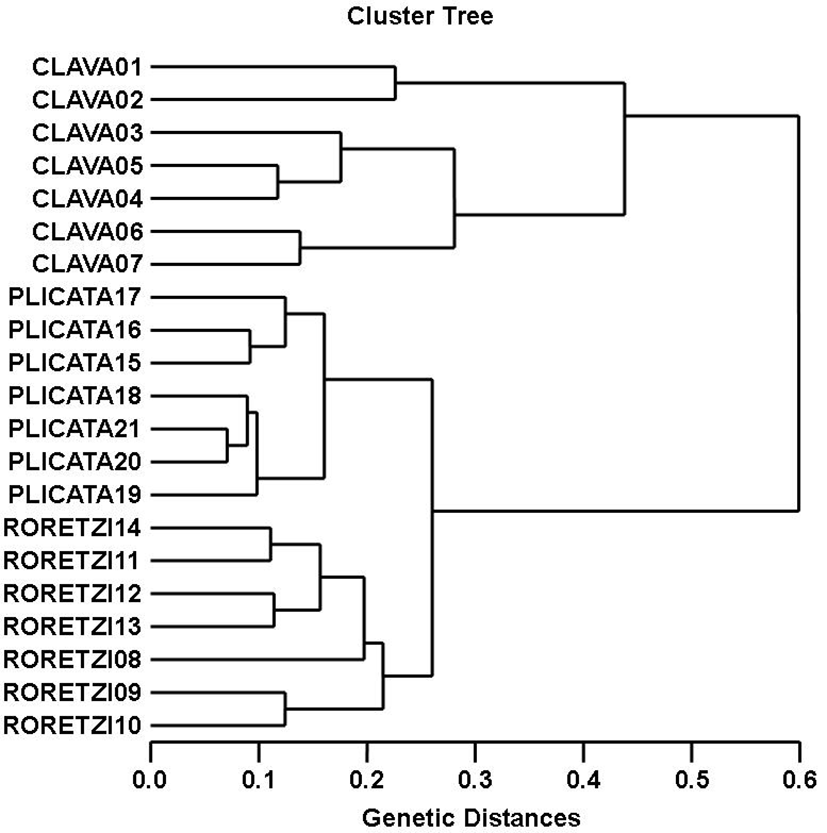
DNA fragments identified in this study may be useful as a DNA marker. This cluster analysis revealed the patterns similar to those posited by Cagigas et al. (1999) and Yoon & Kim (2004). By means of the cluster analysis of genetic similarity values obtained from genetic data, the genetic distance ranged from 0.091 to 0.316 within and among four natural Spanish populations of brown trout (Salmo trutta) (Cagigas et al., 1999). The values of the pairwise comparisons of unbiased genetic distance between the populations of the Indian major carp (Catla catla) from the combined data for the four primers, ranged from 0.025 to 0.052 (Islam et al., 2005). They reported that the Padma and the Jamuna populations were separated from each other with the lowest genetic distance (D=0.025). The genetic distance between the Indian Ocean lobster and the Korean slipper lobster species ranged between 0.040 and 0.612 (Park et al., 2005). They suggested that this genetic technique could be used to discriminate between different river populations of major carp. Consequently, PCR analysis generated on the genetic data displayed that Styela clava (SC) species was widely divided from Halocynthia roretzi (HR) species. Oppositely, individuals of Halocynthia roretzi species were properly closely related to those of Styela plicata species, as revealed in the hierarchical dendrogram of genetic distances. As stated above, the prospective of this analysis to identify pinpointing markers for the identification of the other vertebrates and/or invertebrates species has also been verified (Partis & Wells, 1996; Tassanakajon et al., 1998; Mamuris et al., 1999; McCormack et al., 2000; Diaz-Jaimes & Uribe-Alcocer, 2003; Song & Yoon, 2013). Generally speaking, this PCR analysis been applied to determine specific markers unambiguous to species, genus and geo-graphical population, as well as genetic polymorphism in diverse species of living being (McCormack et al., 2000; Yoon & Kim, 2004; Park et al., 2005; Oh & Yoon, 2014). Preferentially, the habitat classification of the class Ascidiacea is based on morphological variations in tunic shape, tunic color, length and weight as well PCR analysis. In future, further more research is necessary for more profound population/species identification in ascidians.
