INTRODUCTION
The rock bream Oplegnathus fasciatus belongs to the family Oplegnathidae and is widely distributed in the Indian and Pacific Oceans, including Korea, Japan, Taiwan, and Hawaii. Adults inhabit coastal rocky reefs, while juveniles associate with drifting seaweed and feed on zooplankton. It is an economically important resource in the fishing industry in Korea (Oh et al., 2007; Lee et al., 2016). Due to the rapid development of the aquaculture industry in recent years, the spread of bacterial and viral diseases has resulted in economic losses, including massive losses and reductions in productivity (Jung & Oh, 2000; Lee et al., 2016).
Rock bream iridovirus (RSIV) infection has been reported in marine cultured fish in Korea, Japan, China, Taiwan, and Thailand (Inouye et al., 1992; Chua et al., 1994; Kasornchandra & Khongpradit, 1997; Miyata et al., 1997; Jung & Oh, 2000). This virus is known to cause high mortality and result in economic losses (Jung & Oh., 2000). Rock bream infected with RSIV become lethargic, and exhibit severe anemia, enlargement of the spleen, necrosis of the kidney and spleen, and petechiae of the gills (Inouye et al., 1992; Jung et al., 1997; Nakajima & Maeno, 1998).
The immune system is a physiologically important system in the defense against pathogen invasion and to maintain homeostasis in the defense against antigens, external factors that invade organisms. Adaptive immunity is the immune response in which antigen-specific immune cells are activated and react with antigens through specific receptors. The generation of antibodies against a particular pathogen takes a day or several days (Medzhitov & Janeway, 1997; Magnadottir et al., 2006). Moreover, because there is a delay before the initial adaptive immune response takes effect, the innate immune response plays a critical role in the control of infections during this period (Lee et al., 2016).
The type I interferon (IFN) system plays a critical role in innate antiviral immunity in fish (Collet & Secombes, 2002; Schultz et al., 2004; Robertsen, 2006). In this system, direct antiviral responses are carried out by induction of hundreds of IFN-stimulated genes (ISGs) (Sarkar & Sen GC, 2004). The 15 kiloDalton IFN-stimulated gene (ISG15) was the first identified ISG, and forms covalent conjugates with cellular proteins (Farrell et al., 1979; Loeb and Haas, 1994). ISG15 activity occurs through the covalent conjugation (ISGylation) of the LRGG motif, situated at the C-terminal end of all ISG15 proteins, to cellular or viral proteins (Loeb & Haas, 1992; Dao & Zhang, 2005). Extracellular ISG15 displays cytokine-like functions by inducing IFN-g in T-cells and stimulating natural killer cell proliferation (D’Cunha et al., 1996). Overexpression of ISG15 in IFN-a/b receptor knockout mice attenuates Sindbis viral infection and ISGylation was shown to be responsible for this antiviral effect (Lenschow et al., 2005).
Fish ISG15 sequences have been identified in turbot (Scophthalmus maximus), olive flounder (Paralichthys olivaceus), tongue sole (Cynoglossus semilaevis), rock bream (Oplegnathus fasciatus), Northern snakehead (Channa argus), black rockfish (Sebastes schlegelii), Atlantic cod (Gadus morhua) and Atlantic halibut (Hippoglossus hippoglossus). In addition, several ISG15 members have been studied in various defense mechanisms linked to pathogen infection (Seppola et al., 2007; Baeck 2008; Motoshige et al., 2011; Øvergård et al., 2012; Lin et al., 2015; Moreno et al., 2016).
In this study, we examined the expression patterns of ISG15 in several rock bream tissues. In addition, we analyzed the temporal expression profile of RbISG15 in immediate and continuous immune responses following RSIV infection. Our results improve our understanding of the distribution and the functional roles of ISG15 against viral infection.
MATERIALS AND METHODS
Rock bream (Oplegnathus fasciatus) was obtained from the Genetics and Breeding Research Center of the National Institute of Fisheries Science (NIFS, Geoje, Republic of Korea) and maintained under a natural photoperiodin 30-ton tanks. Tissue samples were prepared from various tissues, including brain, eye, gill, intestine, kidney, liver, muscle, spleen, and stomach, obtained from healthy rock bream (total length approximately 19 cm, 2 years old) and stored at –80℃ until required. Whole body samples were also prepared from healthy rock bream (total length approximately 10 cm, 5-6 months old). Deformed and diseased fish were excluded from all experiments. Fish were acclimatized to the experimental conditions for 1 week before the start of the experiment. Tissue collection and pathogen injection were conducted on anesthetized fish and samples were collected under aseptic conditions.
The RSIV challenge experiment was conducted by randomly selecting rock bream specimens, dividing them into challenge and control groups, followed by injection with 100 μL of RSIV suspension (102 TCID50 virus/fish) or phosphate-buffered saline (PBS), respectively (Umasuthan et al., 2013). Each group was maintained at a temperature of 20℃ in a recirculation system, without flow and feeding. Samples from the gill, kidney, liver, muscle, spleen, and whole body were collected at 0, 3, 6, 12, and 72 h post-injection and frozen in liquid nitrogen. All samples were obtained and analyzed in triplicate and samples were ground using a homogenizer for RNA extraction.
Total RNA from the rock bream specimens was extracted using TRI solution (BSK-Bio Co.) according to the manufacturer’s protocol. After extraction of total RNA, DNase-I (Sigma-Aldrich) was used to eliminate genomic DNA contamination. Total RNA was spectrophotometrically (BioTek, Gen 5.2) evaluated for quantity and quality and stored at –80℃ until further use. The isolated RNA was converted into complementary DNA (cDNA) using a Transcriptor First Strand cDNA Synthesis Kit (Roche Ltd., Switzerland). The amplification was performed with Solg™ Taq DNA Polymerase (SolGent Co., Ltd.) in a My Cycler Thermal Cycler (Bio-Rad Laboratories Inc., Hercules, CA, USA) using the following parameters: denaturation at 95℃ for 5 min, followed by 35 cycles of denaturation at 95℃ for 30 s, annealing at 58℃ for 30 s, and elongation at 72℃ for 30 s. The amplified PCR products were analyzed by electrophoresis on a 1.5% (w/v) agarose gel. Specific primers for rock bream ISG15 were designed using the Primer 3 program (Table 1). Relative amounts of each mRNA were quantified by dividing by the density of a housekeeping gene (Cho et al., 2006).
qRT-PCR (ABI 7500, Applied Biosystems) was performed on rock bream samples to quantify the expression of ISG15. The primer pairs used for qRT-PCR amplification of the internal fragments of these genes are listed in Table 1. Isolated RNA was converted into cDNA using a Transcriptor First Strand cDNA Synthesis Kit. RNA was converted to cDNA (100 ng/μL) and used as a template for qRT-PCR. The Fast SYBR Green PCR Master Mix (Applied Biosystems) was used for the reaction, with the following conditions: an initial denaturation step for 20 s at 95℃, 40 cycles of 3 s at 95℃ and 30 s at 60℃, followed by a final dissociation stage. One microliter of cDNA was amplified in a 20-μL reaction using Fast SYBR Green PCR Master Mix with each primer at a final concentration of 500 nmol/L. The 2−ΔΔCt method was used to normalize differences in the amounts of total cDNA in the reaction using actin as an internal standard control (Pfaffl, 2001). The normalized value for ISG15 was then calibrated to the value for the brain or RSIV injection at 0 h, which was assigned a value of 1, by the standard curve method (Applied Biosystems).
The ISG15 mRNA levels were normalized to those of an internal control gene. The statistical analyses of gene expression were performed using one-way analysis of variance. Significant differences from control values were determined at p<0.05 (*) and considered extremely significant at p<0.01 (**). All results are represented as the mean ± standard error of at least three independent replicates.
RESULTS
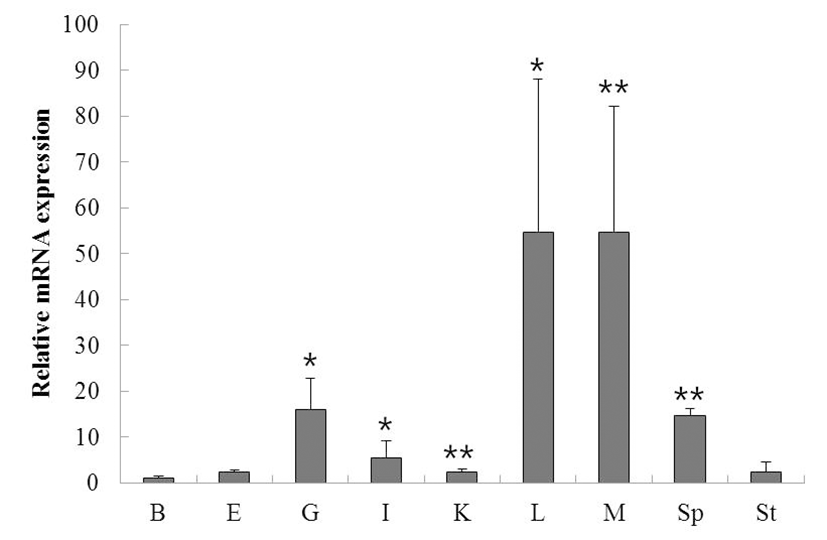
To determine the function and localization of rock bream ISG15, we evaluated the expression of ISG15 in various tissues from healthy rock bream (total length approximately 19 cm, 2 years old) by qRT-PCR. ISG15 was highly expressed in the liver, muscle, spleen, and gill. When standardized to the brain (1.0-fold) of healthy rock bream, ISG15 mRNA levels were increased in the eye (2.4-fold), gill (15.9-fold), intestine (5.4-fold), kidney (2.4-fold), liver (54.7-fold), muscle (54.7-fold), spleen (14.8-fold), and stomach (2.4-fold). These results show that ISG15 mRNA expression differs significantly in different tissues (Fig. 1; supplementary Table 1).
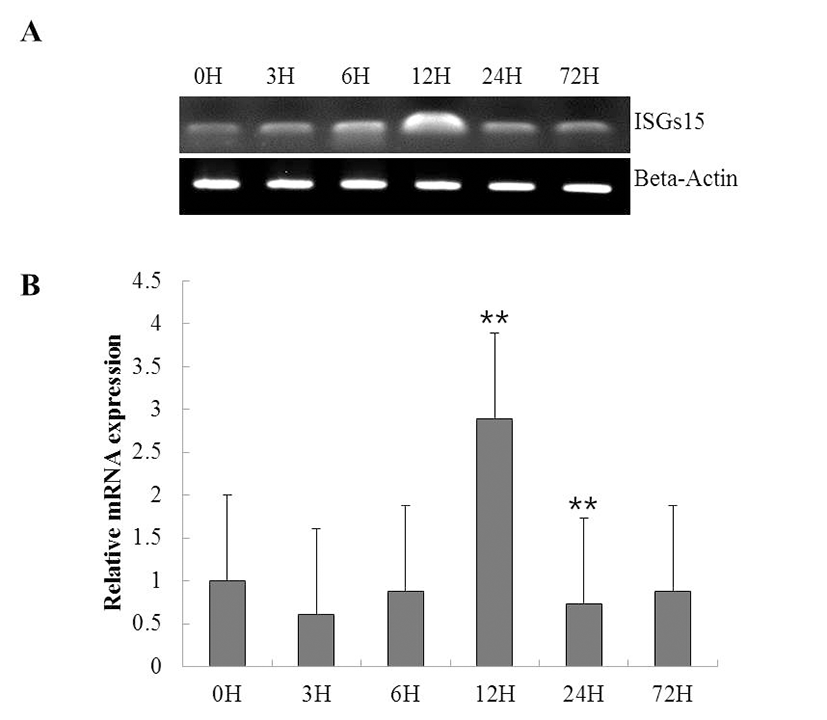
To investigate the function of ISG15 in the immune response to viral infection, we assessed the expression of the gene using RT-PCR in a time course of whole rock bream infected with RSIV. ISG15 was ubiquitously expressed at all time points, but significantly increased at 12 h (Fig. 2A). qRT-PCR was performed to quantify the expression of ISG15 (Fig. 2B). As shown in Fig. 2B, although ISG15 expression remained at almost basal levels until 6 h post-infection (0.8-fold), it increased exponentially at 12 h post-infection (2.8-fold) and was reduced at 24 h post-infection.
Characterization of the temporal patterns of ISG15 mRNA expression in several tissues, including the gill, liver, muscle, and spleen, during pathogen infection was performed via qRT-PCR analysis. Gene expression was normalized to actin and 0 h expression in several tissues and calibrated to the assigned value of 1 by the standard curve method. The real-time PCR data analyzed by the 2−ΔΔCt method indicated that expression challenge with RSIV resulted in a significant increase in ISG15 mRNA expression in several tissues (Fig. 3A-3C). In particular, the highest expression level was detected in the kidney, where ISG15 mRNA levels were significantly induced after 1 h (1.3-fold), peaked at 24 h (123.3-fold), and decreased at 72 h (24.3-fold) (Fig. 3A). In muscle, ISG15 mRNA was significantly induced after 3 h (3.0-fold), peaked at 6 h (15.5-fold), and decreased at 72 h (4.7-fold) (Fig. 3B). Similar to muscle in RSIV-infected rock bream, the expression of ISG15 mRNA was induced following RSIV infection in the spleen (1.5-fold at 3 h post-infection, 3.65-fold at 6 h post-infection, and 1.16-fold at 72 h post-infection) (Fig. 3C). In contrast, the expression levels of ISG15 during RSIV challenge in the gill and liver remained virtually unchanged (data not shown).
| Sample name | CT |
|---|---|
| Brain | 31.021900177 |
| Eye | 31.188400269 |
| Gill | 30.866903305 |
| Intestin | 32.704456329 |
| Kidney | 31.963361740 |
| Liver | 32.925949097 |
| Muscle | 30.991914749 |
| Spleen | 28.597431183 |
| Stomach | 33.255867004 |

DISCUSSION
Rock bream is an economically important aquaculture species, but the damage caused by diseases is increasing. Therefore, to reduce these damages, it will be necessary to understand its immune responses and study its immune system. ISG15 is a small ubiquitin-like protein that plays an important role in mammalian immunity by covalently binding target proteins in a process known as ISGylation (O'Farrell et al., 2002; Schneider et al., 2014). ISG15 has been identified in many species, including zebrafish (Langevin et al., 2013), orange-spotted grouper (Huang et al., 2013), Atlantic cod (Furnes et al., 2009), and tongue sole (Wang et al., 2012), and is induced by IFN. Although ISG15 is known to interfere with viral replication and infectivity to limit intracellular viral infections, the detailed antiviral mechanism is not yet known (Durfee et al., 2010; Poynter et al., 2016).
In this study, we analyzed ISG15 mRNA levels from various healthy tissues in rock bream (Fig. 2). qRT-PCR analysis showed that the ISG15 gene was universally expressed in all tissues. Similarly, Liu et al. (2010) reported that the ISG15 gene can be expressed in all tissues in the red drum, and that the expression level of the ISG15 gene in the liver was the highest among the examined tissues. However, no significant induction of ISG15 expression was observed during RSIV infection, likely because the basal level of ISG15 was enough to cope with the stress induced by RSIV infection.
The expression of ISG15 is influenced by pathogenic infection (Dao & Zhang, 2005; Liu et al., 2010). Previous studies have shown that ISG15 exhibits antiviral activity and can inhibit the production of a wide range of DNA and RNA viruses (Lenschow et al., 2007, Harty et al., 2009). We analyzed the expression levels of ISG15 mRNA in young rock bream following artificial RSIV infection (Fig. 3). RT-PCR analysis indicated that ISG15 was expressed ubiquitously at all times, but significantly increased at 12 h. ISG15 mRNA levels were significantly induced after 6 h, peaked at 12 h, and decreased at 24 h. These results indicate that ISG15 mRNA levels respond quickly to RSIV stimulation (Dao & Zhang., 2005). ISG15 is known to respond quickly to IFN and virus treatments and similar patterns of expression have been reported in several species (Seppola et al., 2007; Kim et al., 2010; Liu et al., 2010; Huang et al., 2013).
Following RSIV infection, rock bream ISG15 expression was acutely increased in the kidney, muscle, and spleen at 6 h and decreased at 72 h post-infection. Expression patterns of ISG15 mRNA following RSIV infection were similar. ISG15 expression in uninfected kidneys of rock bream was low compared to other tissues, but in RSIV-infected tissues, expression was high at 24 h. Based on these results, the changes in the temporal expression of ISG15 in the kidney were induced by RSIV infection. According to Wang et al. (2012), the expression of Cynoglossus semilaevis ISG15 (CSISG15) is increased by microbial pathogens, and CSISG15 is released into the extracellular environment following viral infection. Thus, the CSISG15 protein likely acts as a cytokine to activate various immune cell responses.
These results suggest that ISG15 is regulated by the antiviral immune system. This study contributes to our understanding of the rock bream immune system. The expression pattern of ISG15 differed between the whole body and specific tissues following RSIV infection. Therefore, it will be necessary to study the expression of ISG15 based on growth differences in rock bream.
