INTRODUCTION
Solen corneus is, ecologically warm water bivalve species, belonging to family Solenidae, class Bivalvia, widely distributed on the coast of the Yellow Sea, southern sea and Jeju Island in the Korean Peninsula, the Bo Hai of China and the Hokkaido of Japan. Genomic DNA samples isolated from razor clam (Solen corneus) were obtained from Seocheon of the Yellow Sea in the Korean Peninsula. Under the natural ecosystem, the clams dwell in the tidal flats consisting of a lot of mud, sand and slime in the coastal mud flat where the freshwater is flowed temporarily. Like other clams basically, the rate at which the clam grows depends very much on water quality. The outer shell color of this clam is yellowish and inner shell color is white. Mainly, there are marked differences of the size, color and shape in razor clam along with the ecological circumstances of habitat such as harsh and severe period. In general, there are marked alterations of the shell weight, size, color and shape in Solen corneus in line with the ecological environments of habitat such as prey, rock crystal, water temperature, nourish and hard period.
The razor clam is environmentally and biologically very important bivalves in the Korean Peninsula. Nevertheless, these kinds of Korean bivalve, which are recognized important reproductively (Chung et al., 1986), biochemically (Han et al., 2005; Chung et al., 2006), as well as histologically (Han et al., 2005) are not genetically and/or molecular-biologically studied like other shellfishes. There is a need to understand the genetic characters and conformation of this clam population in order to evaluate precisely the exactly genetic consequence. PCR-based molecular research methods have been applied to investigate the genetic characters of numerous life organisms (Callejas & Ochando, 1998; Muchmore et al., 1998; Zhou et al., 2000; Chenyambuga et al., 2004). Particularly, the specific markers peculiar to the line, the breed, the species, the genus or the geographical populations have been applied for the of individuals and species, hybrid parentage and for the monitoring of DNA markers (Partis & Wells, 1996; Klinbunga et al., 2000; McCormack et al., 2000; Park et al., 2005; Kang & Yoon, 2013; Oh & Yoon, 2014). The author undertook clustering analyses to reveal the Euclidean genetic distances of intra- and between-population of Solen corneus from Seocheon of the Korea, by engaging with the precisely designed oligonucleotide primer sets.
MATERIALS AND METHODS
PCR research was performed on DNA samples extracted from a total of 22 individuals using seven oligonucleotides primers. Foot muscle tissues were collected separately from two Solen corneus clam populations, respectively. DNA extraction should be carried out according to the separation and extraction methods (Yoon & Kim, 2004). 600 µL of chloroform was added to the mixture and then inverted (no phenol). After several rinsings, lysis buffer Ⅰ (155 mM NH4Cl; 10 mM KHCO3; 1 mM EDTA) was added to the samples, and the mixture tubes were gently inverted. Ice-cold 70% ethanol was added, and then the samples were centrifuged at 19,621 g for 5 minutes to extract DNA from the lysates. The concentration of the extracted genomic DNA was measured with the optical density ratio at 260 nm by a spectrophotometer (Beckman Coulter, Buckinghamshire, UK). The DNA pellets were then incubation-dried for more than 12 hours, maintained at -40℃ until needed and then dissolved in the distilled water. PCR was performed using programmable DNA Thermal Cycler (MJ Research Inc., Waltham, MA, USA). Amplification products were separated by electrophoresis in 1.4% agarose gels with TBE, using 100 bp DNA ladder (Bioneer, Daejeon, Korea) as DNA molecular weight marker and detected by staining with ethidium bromide (Song & Yoon, 2013). The electrophoresed agarose gels were illuminated by ultraviolet rays, and photographed using a photoman direct copy system (PECA Products, Beloit, WI, USA). Seven oligonucleotides primers, BION-44 (5’-TGCCGAGCTG-3’), BION-51 (5’-AACGCGCAAC-3’), BION-58 (5’-GCCTG TGTC- 3’), BION-63 (5’-CCAGACAAGC-3’), BION-71 (5’-AGGGTTCGGT-3’), BION-77 (5’-TAGAGACTCC-3’), and BION-82 (5’-AAGGATCAGA-3’) were shown to generate the unique shared loci to each clam population and shared loci by the two clam populations which could be clearly counted. Similarity matrix including bandsharing values between different individuals in the two razor clam populations was generated allowing formula of Jeffreys & Morton (1987) and Yoke-Kqueen & Radu (2006). A clustering tree was assembled using similarity matrices to yield a hierarchical dendrogram, which was aided by the Systat version 10 (SPSS Inc., Chicago, IL, USA).
RESULTS AND DISCUSSION
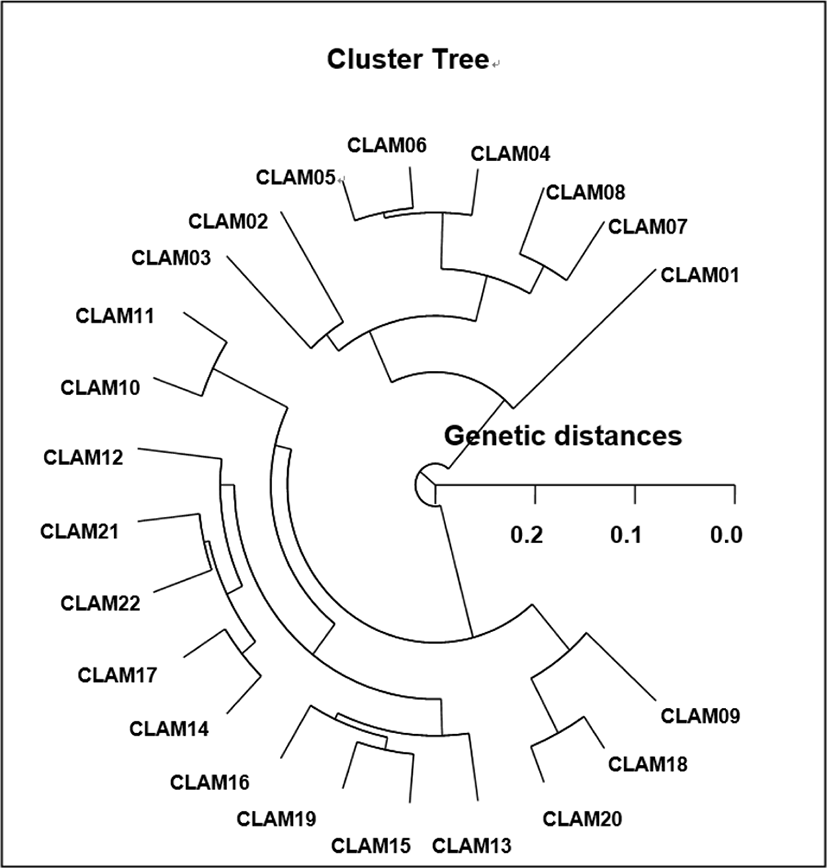
The seven selected oligonucleotides primers BION-44 BION-51, BION-58, BION-63, BION-71, BION-77 and BION-82 generated the total number of loci, average number of loci per lane and specific loci in two razor clam populations, as illustrated in Table 1. Here, the complexity of the banding patterns varied dramatically between the primers from the two clam populations, as revealed on Fig. 1.
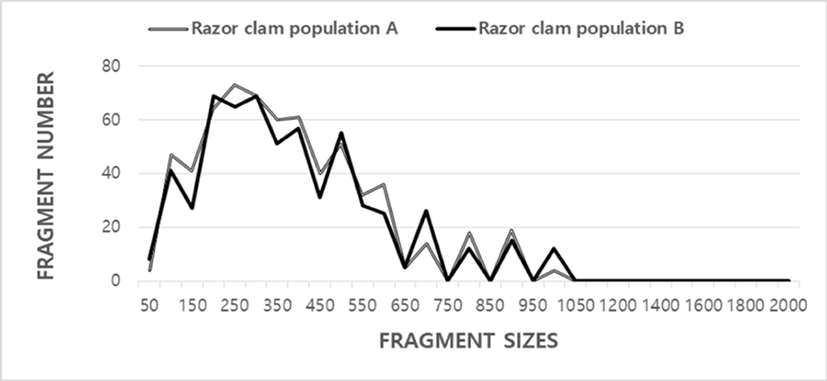
Seven oligonucleotides decamers were used producing a total of 639 scorable bands in population A and 595 in population B, respectively, ranging in size of DNA fragments from larger than approximately 50 to less than 1,100 bp, as illustrated in Table 1. Eighty fragments ranging from 200 bp to 2,200 bp were unambiguously counted in the black tiger shrimp (Penaeus monodon) (Tassanakajon et al., 1998). The DNA fragments obtained using the four primers ranged from 100 to 2,300 bp in the brittle star (Amphiura filiformis) (McCormack et al., 2000). In the present study, seven oligonucleotides decamers generated 39 specific fragments (6.10%) in population A and 47 (7.90%) in population B, respectively (Table 1). The primer BION-58 generated 66 unique loci to each population, which were determining each population, approximately 100 bp, 150 bp, 250 bp, 350 bp, 400 bp and 500 bp, in the population A, as illustrated in Table 2. Extraordinarily, the primer BION-77 detected 22 shared loci by the two clam populations, major and/or minor fragments of sizes 250 bp, which were equivalent in all samples. With regard to average bandsharing value (BS) results, individuals from population A (0.799) exhibited higher bandsharing values than did individuals from population B (0.771) (Table 3). In the present study, the dendrogram obtained by the seven oligonucleotides primers indicates two genetic clusters: cluster 1 (CLAM 01, 02, 03, 04, 05, 06, 07, 08, and 11) and cluster 2 (CLAM 09, 10, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, and 22) (Fig. 2). Among the twenty-two clams, the shortest genetic distance that displayed significant molecular differences was between individuals clam no. 20 and no. 18 from the cluster II (genetic distance=0.036), while the longest genetic distance among the twenty-two individuals that exhibited significant molecular differences was between individuals clam no. 08 and clam no. 12 (genetic distance=0.280). Comparatively, individuals of cluster I were greatly closely related to that of cluster II, as shown in the hierarchical dendrogram of Euclidean genetic distances. The genetic distance between individuals approved the existence of close relatedness in the cluster II. Above-mentioned, a dendrogram discovered close relationships between individual structures within three geographical bivalve populations (McCormack et al., 2000). Cluster analysis showed a similar pattern to that illustrated by Oh & Yoon (2014). They reported that cluster analysis, which indicated three genetic clusters, and the dendrogram revealed a close relationship between the individual identities within two species. In the other invertebrates, cluster analysis of the pairwise population matrix, generated from genetic data, showed that geographically close populations be inclined to cluster together in the blacklip abalone (Huang et al., 2000). Two S. corneus populations can be evidently discriminated by PCR-based techniques. The potential of oligonucleotides amplified polymorphic DNAs to discover diagnostic markers for line, breed, species, genius and population identification in crustaceans (Partis & Wells, 1996; Callejas & Ochando, 1998; Huang et al., 2000; Klinbunga et al., 2000; McCormack et al., 2000; Kim et al., 2000; Park et al., 2005; Oh & Yoon, 2014) has also been well recognized. Ultimately, PCR analysis has exposed the significant genetic distance among two razor clam populations. PCR fragments revealed of in this study may be valuable as a DNA marker the three regional populations to differentiate. Overall, the population grouping of S. corneus is founded on morphological variations in shell size, shell weight, shell type, shell color, feet length, mantle edge and etc. It is implicated that differences in such traits reflect exclusive origins or genetic identity (Chenyambuga et al., 2004). High levels of a significant genetic distance among two razor clam populations showed this PCR method is one of the most suitable tools for individuals and/or populations biological DNA researches (Muchmore et al., 1998; Tassanakajon et al., 1998; Zhou et al., 2000; Yoon & Park, 2002; Yoon & Kim, 2004; Song & Yoon, 2013). Consequently, this procedure can also be applied to other species of Solenidae and create technically-convenient the analysis of many samples in a little while.
| Population | Population A | Population B |
|---|---|---|
| Population A | 0.799±0.010a | 0.734±0.005b |
| Population B | - | 0.771±0.009a |
Each value is a result of three different experiments.