INTRODUCTION
Culture of eel Anguilla japonica totally depends on glass eel collected from the wild despites of the long time effort for more than 50 years to produce them artificially. Artificial production of glass eel has been succeeded in some laboratories of Japan (Tanaka et al., 2003) and Korea (Kim et al., 2013, 2014). However, survival rates of embryo and larvae are extremely low and still far from industrial scale. Large quantity of eggs in this species fail to be fertilized and many of fertilized eggs are unable to hatch. Larvae of eel absorb egg yolk up to 8 days after hatching but the majority of hatched larvae die before they reach the stage of first feeding (Ohkubo et al., 2008), indicating the importance of yolk processing in this species.
Many previous studies have reported that survival of fish embryos during early development could be associated with yolk processing by several proteases (Carnevali et al., 2001; Tingaud-Sequeira et al., 2011; Palomino et al., 2017). The key enzymes known to be involved in the process are cathepsins and lipoprotein lipase. Cathepsin family can be divided several subgroups accoding to the amino acids that degrade, including typical cysteine protease, cathepsin B and L, and an aspartate protease, cathepsin D (Duve, 1983; Roberts, 2005; Liaudet-Coopman et al., 2006). Recently, Palomino et al. (2017) showed that cathepsin can determine egg quality of a marine fish species. In yellowtail kingfish Seriola lalandi, mRNA expressions and enzyme activities of cathepsin B, cathepsin D and cathepsin L were associated with the survival of embryos during the early development (Palomino et al., 2017). In addition, during maturation, the key enzymes such as cathepsin B, D, and L were involved in the egg development and early embryonic development of fish (Carnevali et al., 1999a, 1999b, 2001, 2008; Kwon et al., 2001; Tingaud-Sequeira et al., 2011).
These enzymes seem to play the role by working on the hydrolysis process of egg yolk protein during embryonic development (Tingaud-Sequeira et al., 2011; Palomino et al., 2017). Transcripts of cathepsin B and D are produced and stored in oocytes during vitellogenesis in fish (Carnevali et al., 1993; Matsubara & Sawano 1995). After fertilization, the transcripts stored in the eggs are translated into enzymes that hydrolyze the stored yolk protein and supplies the energy and basic material needed for early development (Sire et al., 1994; Carnevali et al., 1999; Kwon et al., 2001; Hiramatsu et al., 2002; Tingaud et al., 2011). However, despite the importance of egg yolk processing, information on this process in eggs of eel is very limited.
A study has been reported on the importance of buoyancy (i.e., floating or sinking) of eggs in Japanese eel as a criterion for evaluating the quality of eggs (Unuma et al., 2005). In this study, we investigated differences in the expression of yolk processing enzyme genes between floating eggs and sinking eggs, and also the relationship between the gene expressions of the enzymes and fertilization rates in the fertilized eggs of this species.
MATERIALS AND METHODS
Eels were purchased from a private eel farm in Gochang, Korea (females 550–700 g, males < 500 g in body weight, b.w.) They were acclimated for 2 weeks at 20±1℃ without feeding in a 2,000 L indoor tank using filtered seawater before the beginning of hormone treatment for the induction of sexual maturation. Males were treated with human chorionic gonadotropin (HCG; Daesung microbiological labs) at the dose of 1 IU/kg, b.w. Female eels were treated with salmon pituitary extract (SPE) at a dose of 20 mg/fish, once a week. After 10 to 15 injections, fish were measured for body weight. When fish were ready to spawn, 170 μL of 17, 20β-dihydroxy-4-pregnen-3-one (DHP) was injected to each female in order to induce ovulation. Fish spawned about 15 hours after the injection. The eggs were artificially fertilized with sperm, and fertilization rate was examined.
Fertilization eggs from each female were collected separately into 15 mL conical tubes after dividing the floating eggs and sinking eggs. All sampling procedure took less than an hour after fertilization. Eggs were stored at -80℃ until extraction. Total RNA was extracted from the stored eggs using TRIzol(r) Reagent (ambion, USA). Extracted total RNA was quantified using nanodrop-2000 (Thermo, USA) and used to synthesize cDNA.
Transcript levels of cathepsin B gene (ctsb), cathepsin D gene (ctsd), cathepsin L gene (ctsl) and lipoprotein lipase gene (lpl) in the fertilized eggs of eel was investigated by real-time PCR (RT-qPCR) using cDNAs of floating or sinking eggs with different fertilization rate as templates. Primers for each gene were designed using Primer 3 software (version 2.2.3) (Table 1). Each RNA sample (1 μg) was revese transcribed using TOPscrit(tm) RT Dry MIX (Enzynomics, Korea). For RT-qPCR, 10 μL of 2X Pre-Mix SY BR Green (Enzynomics, Korea), 1 μL of each primer, 5 μL of cDNA template (50-fold dilution) and 3 μL of nuclease free water (NFW) were used. Sequence information of ctsb, ctsd, ctsl, lpl of eel used in the experiment was found in online databases http://molas.iis.sinica.edu.tw/jpeel/ and National Center for Biotechnology information (www.ncbi.nlm.gov).
Data were presented as mean±SEM. Statistical difference of the expression levels between floating eggs and sinking eggs was analyzed by one-way ANOVA and independent two-sample t-test (p<0.05). The relationship between the expressions of yolk processing enzyme genes (ctsb, ctsd, ctsl, lpl) and fertilization rate were shown by regression analysis. Statistical analyses were preformed using SPSS version 18.0.
RESULTS AND DISCUSSION
Expressions of yolk processing enzyme genes did not show significant difference between floating and sinking egg groups in Japanese eel in this study (Fig. 1). This result contradicts the results from other fish species. In gilthead seabream Sparus aurata, cathepsin D activity was higher in sinking eggs than in floating eggs (Carnevali et al., 1999). In European seabass Dicentrarchus labrax, cathepsin D gene was expressed significantly higher level in the sinking eggs, while cathepsin L gene showed a higher expression in the floating eggs (Carnevali et al., 2001).
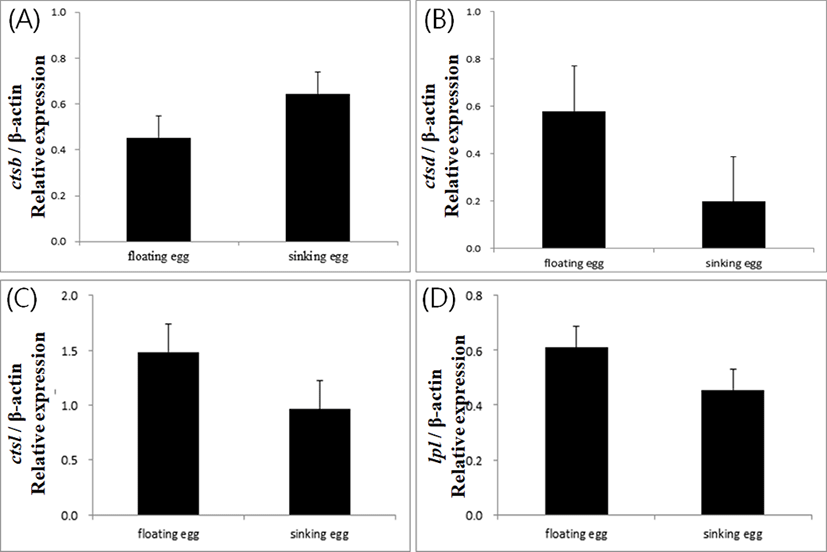
In general, buoyancy of normal healthy eggs differs from species to species in fish. Eggs from salmonid fish are demersal. Sinking eggs are considered to be good in this kind of fish. Eggs from seabream, seabass and many marine fish species are buoyant, and floating eggs with low expression of cathepsin D are considered to be good. In this study, we assumed the floating eggs as good quality and sinking eggs as bad quality, based on the suggestion from Unuma et al (2005). It was expected that the floating eggs of eel would show low expression of ctsd and high ctsl. This discrepancy between the results and expectation might indicate that the mechanism of buoyancy acquirement in eel eggs could differ from the mechanism in other marine fish species. This potential difference of the mechanism could be attributed to the catadromous characteristics of eel.
Another possibility for the unexpected results could be the potential existence of tissue specific transcript variants for the enzymes. The sequences used to design the primers for each enzymes were not originated from ovarian or egg cDNA library. It is possible that egg specific transcripts for yolk processing enzymes would have shown clear differential expression between floating and sinking eggs if there were tissue specific isoforms for each enzyme gene.
Fertilization rates of eggs from 5 different females were different to each other, ranging from 36.0% to 76.0%. Expression level of ctsb decreased significantly as the fertilization rate increased (p<0.05) (Fig. 2A). However, expression levels of ctsd and ctsl did not show significant any relationships with fertilization rate (p>0.05) (Fig. 2B & 2c). Expression level of lpl tended to increase as fertilization rate increases, but showing no statistical significance (p>0.05) (Fig. 2D). In support of this result, the level of ctsb has been suggested to be a criterion for egg quality in yellowtail kingfish and red spotted grouper (Gwon et al., 2017; Palomino et al., 2017). However, they focused on the difference between developmental stages while, in this study, we have looked into differences between egg groups with different fertilization rate. This could mean that our result tells more directly the quality of eggs. Thus, the level of ctsb transcript in eggs could be considered as a potential marker for evaluating egg quality, but not the level of ctsd, ctsl and lpl in this species. High fertilization rate, however, does not always result in high hatching rate and survival rate at the post-embryonic stage. Further study should investigate the relationship between yolk processing enzymes and hatching rate or post-embryonic survival.
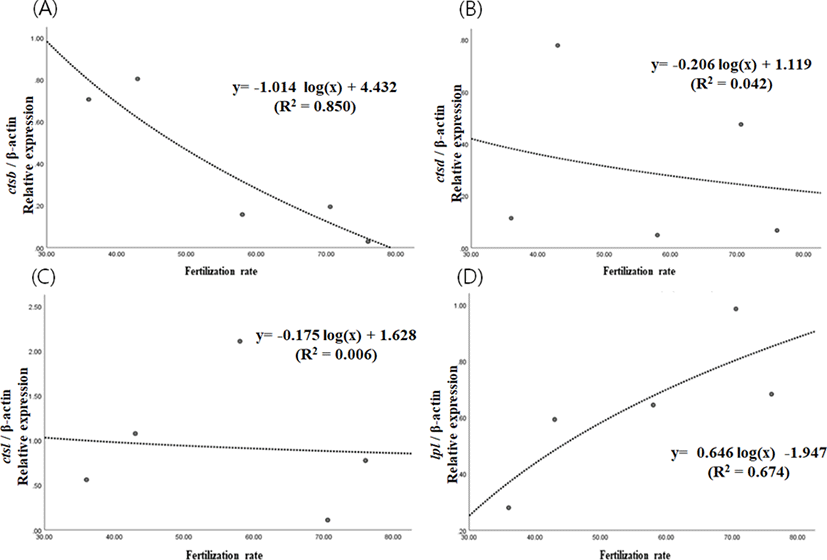
In summary, these results suggest that ctsb expression could be an indicator of egg quality, and that some proteins prone to be digested by cathepsin B could be very important in the process of fertilization and normal cleavage in this species. Concentration of the proteins would be high leading to successful fertilization when the activity of cathepsin B is low in the eggs of the eel. Further study should identify these critical proteins to improve our understanding on the quality of fish eggs. In further studies, other enzymes which are also involved in yolk processing at early embryonic stages need to be investigated. In addition, possibility of tissue specific splicing for yolk processing enzyme genes should be investigated.
