INTRODUCTION
The olive flounder (Paralichthys olivaceus) is widely distributed around East Asian shores and is one of the most economically important marine organisms as it has a 40% share in local sea water fish culture. However, the survival rate of cultured olive flounders is seriously influenced by a variety of diseases, radical changes in temperature, and high-density cultures. In particular, fish in the larval phase are immediately exposed to various microorganisms, hence study on their effective immune system and diseases is a very important prerequisite for the improvement of survival rate (Zapata et al., 1997).
Viral hemorrhagic septicemia virus (VHSV) is known to cause mass mortality in olive flounders (Paralichthys olivaceus), rainbow trouts (Oncorhynchus mykiss), turbots (Scophthalmus maximus), etc. (Kim et al., 2009; Pinheiro et al., 2016; Pereiro et al., 2016). The spleens of infected fish are bloated, full of ascites, and cause extensive bleeding in both internal and external organs including the fins, gills, and eyes (Isshiki et al., 2001; Duesund et al., 2010). Moreover, it has a rapid infection speed and occurs frequently in locally cultured olive flounders (Kim et al., 2003).
The immune system is a biologically important system that blocks the invasion of pathogens, maintains homeostasis, and protects the organism from antigens. In the case of fish, they live in an aqueous environment from the fry stage and depend on their congenital immune systems for survival. The congenital immune system is the first line of protection against various microorganisms and is essential for controlling pathogenic infection (Beck & Habicht, 1996; Fischer et al., 2013).
Chemokine, a main chemoattractant of cytokine that is concerned with congenital immunity, plays the important role of directing the movement and activation of leukocyte across the entire body (Luster, 1998; Mantovani, 1999). Chemokine is divided into CC, CXC, CX3C, and C-chemokine according to the composition of NH2-terminal cys residue (Hsu et al., 2013). In addition, in the immune system, chemokine may be divided into homeostatic chemokine and inflammatory chemokine. In immune reactions, chemokine generally is involved in attracting immunocytes to the damaged region and removing pathogens through phagocytosis.
Most chemokine belongs to the CC and CXC chemokine groups (Murphy et al., 2000). CC chemokine has 2 close cysteines near the terminal of the amino acid and induces the movement of monocytes including NK cells, dendritic cells, and other types of cells. In addition, it is a strong chemoattractant of monocytes, lymphocytes, basophils, and eosinophils (Miller & Krangel, 1992; Taub & Oppenheim, 1993; Murphy, 1994). CXC chemokine has 2 cysteines and another amino acid that is expressed as “X” in the N-terminal, and in the case of mammals, it is classified according to the existence of glutamic acid (E) – leucine (L) – arginine (R) (ELR motif) directly before the first cysteine (Oppenheim et al., 2001). Specifically, a positive ELR motif induces the movement of neutrocytes and acts as coaction of the chemokine receptors CXCR1 and CXCR2. Moreover, it is interleukin-8 (IL-8) that induces neutrocytes to drop into blood flow and enter surrounding tissues. Negative ELR motif, such as CXCL13, has chemical attractancy to lymphocytes (Bazan et al., 1997; Pan et al., 1997).
Chemokine in modern fish is reported in Atlantic cod (Gadus morhua), Atlantic salmon (Salmo salar), channel catfish (Ictalurus punctatus), large yellow croakers (Larimichthys crocea), olive flounders (Paralichthys olivaceus), rainbow trout (Oncorhynchus mykiss), rock breams (Oplegnathus fasciatus), zebrafish (Danio rerio) and others, however, information on immune reaction is limited (Bao et al., 2006; Borza et al., 2010; Grimholt et al., 2015; Khattiya et al., 2004; Kim et al., 2013; Knaut et al., 2003; Semple et al., 2018; Zhou et al., 2018). Therefore, in this study, in order to examine the immune system of fish, analysis was performed on the gene expression of CC and CXC chemokines at different stages and in different tissues of olive founders during the initial occurrence stage and after VHSV infection in the gills, spleen, and kidneys.
MATERIALS AND METHODS
Olive flounders used as test subjects were bred in a 5 ton round water tank at a water temperature of 19±1° with a light period of 15 hours and a dark period of 9 hours at the Breed Research Center of the National Fisheries Research and Development Institute. To observe gene expression at different stages of development, samples taken from test subject up to 18 days after hatching were put in Trizol Reagent (Invitrogen) and stored at −80° until RNA separation. To analyze gene expression in different tissues, the brain, eyes, fins, gills, intestines, kidneys, liver, muscles, spleen, and stomach were extracted from a healthy 8-month-old olive flounder (total length of approximately 30 cm) and stored at −80° until testing.
Artificial VHSV infection testing was performed on healthy olive flounders (total length of approximately 30 cm, 8 months old) by dividing them into control and test groups. The test subjects were moved to a 3 ton round water tank and not fed on the day of or the day prior to inoculation. The test was performed at a water temperature of 13° with a light period of 15 hours and a dark period of 9 hours. The control group was inoculated with 100 μL of phosphate buffered saline (PBS) solution and the test group was inoculated with a VHSV suspension (104.8 TCID50 virus/ fish) (Kong et al., 2009). Sampling was performed from the gills, spleen, and kidneys of olive flounders 0, 1, 6, 12, and 24 hours after inoculation. Stress on test subject was minimized through anesthesia using MS-222 (Sigma, USA) at a concentration of 150 ppm (Noh et al., 2017) and each tissue was stored at −80° until testing.
Total RNA was extracted from test olive flounders using a TRI solution (BSK-Bio Co.). Genomic DNA contamination of the extracted RNA was removed using DNase-I and qualitative and quantitative quantities were measured using a spectrophotometer (BioTek, Gen5.2). The cDNA of the total RNA was synthesized using oligo-d (T)18 primer and a Transcriptor First Strand cDNA Synthesis Kit (Roche Ltd., Switzerland), and qRT-PCR (ABI 7500, Applied Biosystems) was performed under the following conditions to confirm the gene expression of CC and CXC chemokine in the olive flounders (Lee et al., 2018). After performing the initial denaturation process for 20 seconds at 95°, annealing and elongation were performed for 30 seconds at 58° and 60° respectively. 18S rRNA was used as the internal standard control and the gene expression ratio of 18S rRNA was quantified using the 2-ΔΔCt method. In addition, Primer3 CC chemokine, CXC chemokine, and 18S rRNA primer of the olive flounders were designed using the Primer3 program and are shown on Table 1.
All samples were performed three times for accuracy and all data were expressed in average±standard deviation (n=3). Significant differences in data value were checked via one-way ANOVA. Significance testing of the gene expression between each entity was performed using Tukey's HSD test at p<0.05.
RESULTS
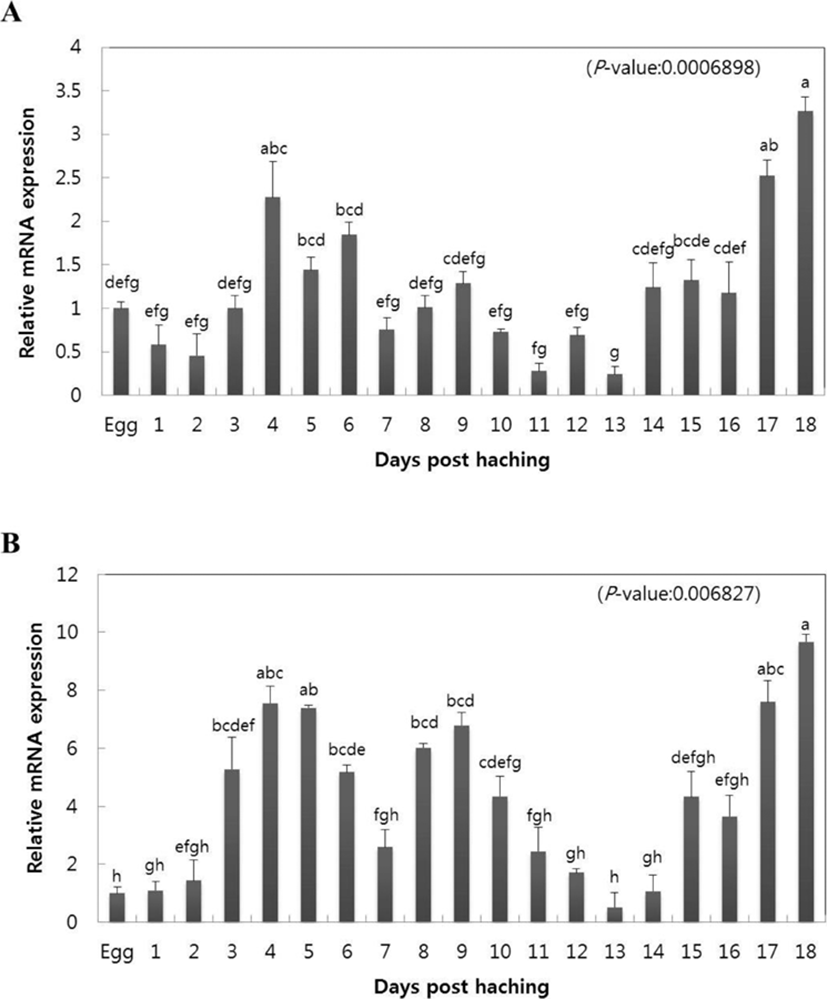
To investigate the congenital immune reaction of olive flounders, the gene expression of CC chemokine, CXC chemokine, and mRNA were analyzed beginning from the initial occurrence stage in olive flounders until 18 days after hatching. Quantitative analysis was performed on CC and CXC chemokines of olive flounders by qRT-PCR using gene specific primer. Relative gene expression of mRNA at each occurrence stage was calculated using the olive flounder reference gene 18S rRNA, and relative tissue specific gene expression was investigated by comparing the results with the eggs. CC chemokine increased at day 4 after hatching (2.27-fold), began to slowly decrease starting from day 5 (1.44-fold), and reached minimum value at day 13 (0.27-fold). Subsequently, the value began to slowly increase starting on day 14 (1.23-fold) and reached maximum gene expression at day 18 (3.26-fold) (Fig. 1A). CXC chemokine started increasing from day 1 after hatching (1.1-fold) and continued to increase until day 4 (7.55-fold). The value subsequently slowly decreased until day 7 (2.59-fold) and reverted to increase until day 9 (6.78-fold). Then, identical to CC chemokine, the value of CXC chemokine showed minimum gene expression on day 13 (0.52-fold) and started to slowly increase after day 14 (1.07-fold), reaching maximum gene expression on day 18 (9.67-fold).
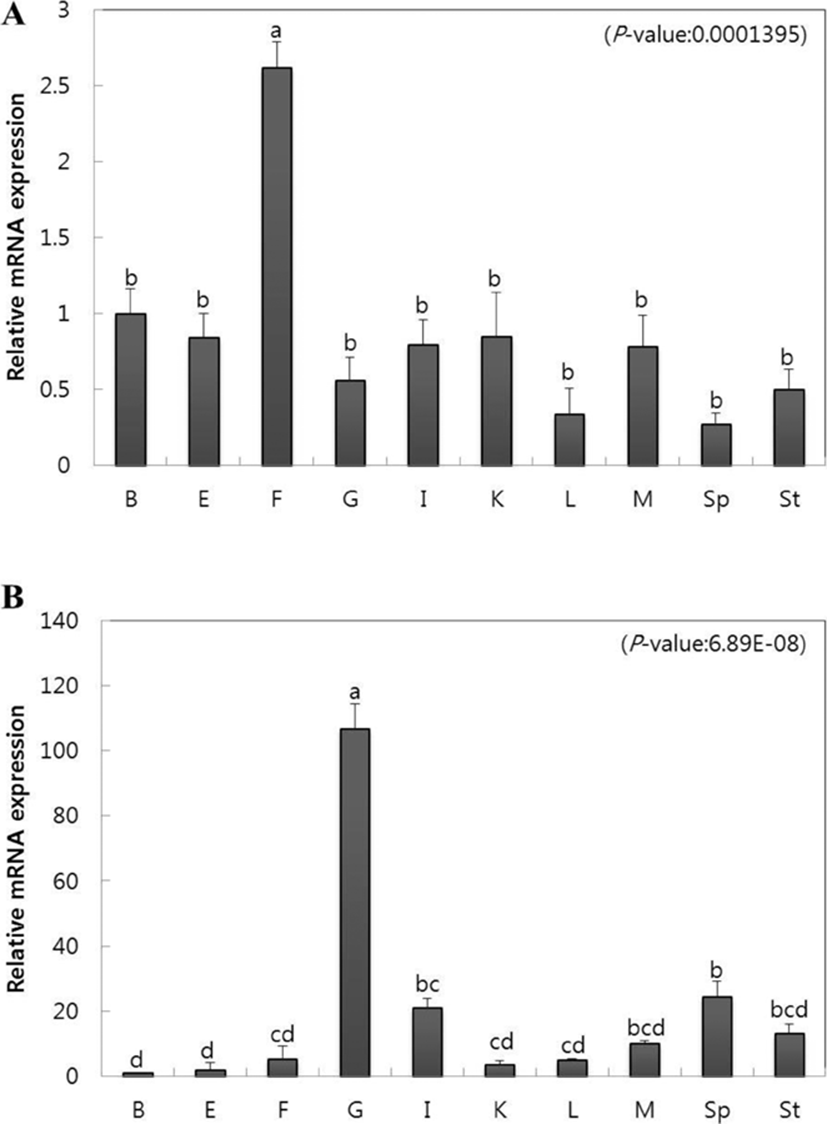
To observe the gene expression of CC and CXC chemokine in tissue, we extracted brain, eye, fin, gill, intestine, kidney, liver, muscle, spleen, and stomach tissues from healthy olive flounders and investigated gene expression using qRT-PCR. Relative mRNA gene expression was calculated for each using olive flounder reference gene 18S rRNA, and relative tissue specific gene expression was investigated by comparing the results with brain tissue. CC chemokine showed the highest gene expression in the fin (2.6-fold) compared to other tissues and showed relatively low gene expression in the liver (0.3-fold) and spleen (0.2-fold) (Fig. 2A). CXC chemokine showed both the highest and lowest gene expression in the gills (106.6-fold) and brain (1-fold), respectively (Fig. 2B). Furthermore, the spleen (24.3-fold), intestine (21.2-fold), stomach (13.1-fold), and muscle (10.2-fold) also showed high gene expression (Fig. 2B).
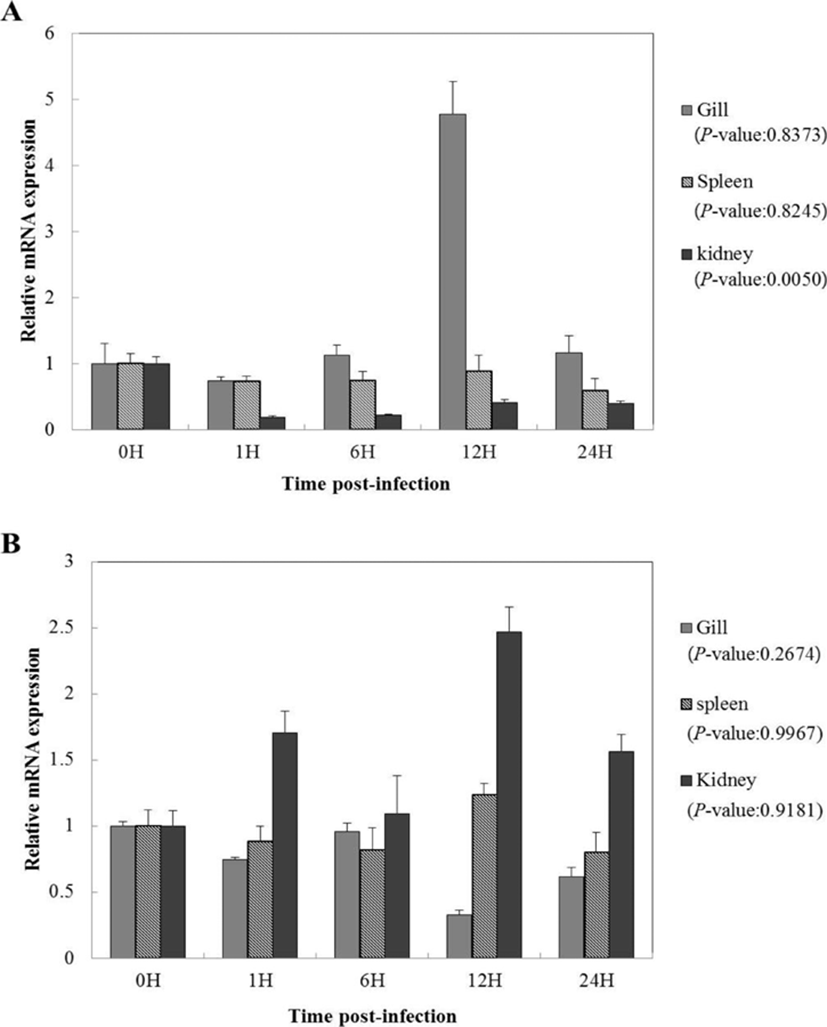
To study the functions of CC and CXC chemokine on immune reaction against viral infection, the gene expression of olive flounders artificially infected with VHSV was recorded according to time elapsed. Relative mRNA gene expression for each time period was calculated using olive flounder reference gene 18S rRNA and relative tissue specific gene expression was investigated by comparing the results with those from 0 hours, the starting point of artificial infection. CC chemokine showed high gene expression in the gills at 12 hours after inoculation (4.7-fold), and in the kidneys, it showed a tendency to decrease after 0 hours (1-fold) (Fig. 3A). Moreover, in the case of the spleen, no overall significant difference was seen (Fig. 3A). CXC chemokine in the gills did not show a large difference from 0 hours to 6 hours after inoculation of the virus and showed a tendency to decrease starting from 12 hours (0.5-fold) (Fig. 3B). In the case of the kidneys, high gene expression was seen at 12 hours (2.4-fold). In the spleen, it showed significant difference from 0 hours (1-fold) to 12 hours (1.2-fold) and started to decrease slowly from day 1 (0.7-fold) (Fig. 3B). CC chemokine after artificial VHSV infection showed high gene expression in the gills, while CXC chemokine showed high gene expression in the kidneys (Fig. 3).
DISCUSSION
Fish, unlike other vertebrates, live in an aquatic environment from their egg stage and are therefore strongly exposed to various pathogens from the fry stage. Their immune system, such as their marrow and lymphatic glands, are not as well evolved as mammals, however, antibodies are produced and microorganisms are destroyed through congenital immune reactions (Magnadottir, 2006). According to Kim (2009), the larvae of olive flounders develop primordial finfolds immediately after hatching, the mouth and anus open at day 4, and the gills open at day 6. In addition, the digestive canal expands towards the stomach and is connected to intestines while the stomach starts to develop and the larvae beings planktonic life. It is reported that the digestive canal becomes completely divided into the esophagus, foregut, midgut, and hindgut by day 14 while the digestive canal continues overall development alongside the growth of body, and that the liver grows in size significantly by day 18. Therefore, when comparing with the results of this study (Fig. 1), it can be seen that the gene expression of CC chemokine and CXC chemokine increases at day 4 when the mouth and anus open and that the maximum gene expression is shown when the digestive canal is divided and the body grows significantly. The results show that CC chemokine and CXC chemokine manifest beginning at the egg stage through to the late stage larval stage, and it can be seen that gene expression increased during the late stage where immune tissues are formed rather than during the initial occurrence stage.
CC chemokine is known to play an important role as a signal material, namely as a chemical chemoattractant, that can attract certain immunocytes, (Xu et al., 2011). Looking at the gene expression of CC chemokine in the tissues of various fish, rock breams (Oplegnathus fasciatus), large yellow croakers (Pseudosciaena crocea), and trout (Scophthalmus maximus) showed high gene expression in the spleen, kidneys, and intestine, while miiuy croakers (Miichthys miiuy) showed high gene expression in the fins (Zhang & Chen, 2008; Chen et al., 2010; Xu et al., 2011; Kim et al., 2013). In addition, in the same manner with CC chemokine in most fish, it can be predicted that in this test (Fig. 2A), CC chemokine manifested in a variety of tissues to provide a homeostatic function through immunosurveillance rather than to function as chemoattractant in immune- related tissues. CXC chemokine, like in other fish, manifested in all tissues (Fig. 2B) and showed high gene expression in the gills, spleen, and kidneys, similar to large yellow croakers (Tian et al., 2010) and rock breams (Oplegnathus fasciatus). However, the degree of gene expression contrasted with this test which implies that gene expression of CXC chemokine in fish tissues may vary by species.
The transient changes in CC and CXC chemokine was analyzed using various inflammatory mediators, and the results are known to play an important role, mainly in the immune response (Kim 2012; Hsu et al., 2013; Kim et al., 2013; Zhou et al., 2018; Semple et al., 2018). Therefore, we aimed to observe the immune response of CC and CXC chemokine through an artificial VHSV infection. AS result of performing subtractive suppressive hybridization using rainbow trout infected with VHSV, CXC chemokines play an essential role in virus defense (Liston & McColl, 2003; Farrell et al., 2002). The kidneys have abundant leukocytes and are an immune-related organ with various immunocytes (Uribe et al., 2011). Therefore, it is thought that the high expression of CXC chemokine as the time elapses in the kidney plays an important role in the immune response of fish (Fig. 3B). However, it is deemed that high gene expression of CC chemokine in the gills is due to exposure to the aquatic environment because, as a respiratory organ that is in charge of discharge and osmoregulation, it is an invasion pathway for countless microorganisms (Fig. 3A; Kim, 2012). Whether these results are related to the role of immune defense requires further investigation.
In this study, analysis of the gene expression of CC and CXC chemokine was performed to examine the immune reaction of fish, and as a result, it is thought that both the CC and CXC chemokines play an important role in the congenital immune system of fish. It is hoped that this study will become valuable baseline data in the study of immune reaction of fish in the future. Additional study on the CC and CXC chemokines in post larval phases where immune tissues are completely formed shall be necessary.
