INTRODUCTION
The olive flounder, Paralichthys olivaceus, is an important marine fish species for aquaculture in Asian countries including Korea, Japan, and China (Kim et al., 2014). It is cultivated mainly on the southern coast and Jeju Island in Korea, with 37,267 tons produced in 2018, worth approximately 422 million USD, or 14.9% of the total aquaculture value in the Korea (http://kosis.kr/statHtml/statHtml.do?orgId=101&tblId=DT_1EW0001&conn_path=I2). Its genome and transcriptome were sequenced as a model for studying asymmetric development during flatfish metamorphosis (Shao et al., 2017).
Actins are essential cytoskeletal proteins in eukaryotic cells that play an important role in cell motility, structure, and integrity (Dominguez et al., 2011). Of the three major vertebrate actin groups, alpha-actins are specific for striated muscle, and cardiac muscle alpha actin 1 (ACTC1) is expressed in cardiac and skeletal muscle (Krasnov et al., 2003; Moll et al., 2006). The alpha-actin promoter has been a critical model for the identification of muscle-specific regulatory elements in vertebrates from fish to mammals (Sartorelli et al., 1990; Higashijima et al., 1997; Krasnov et al., 2003; Moll et al., 2006). The promoter region of the human cardiac alpha-actin (HCA) gene contains transcription factor-binding sites for MyoD, CArG box binding factor, and Sp1 upstream of the TATA box, which are required for muscle-specific expression (Gustafson & Keds, 1989; Sartorelli et al., 1990).
The first fish alpha-actin gene was identified in the pufferfish, Fugu rubripes (Venkatesh et al., 1996). Alpha-actin promoters have been cloned and characterized in several fish species, including zebrafish Danio rerio, medaka Oryzias latipes, and channel catfish Ictalurus punctatus (Higashijima et al., 1997; Kusakabe et al., 1999; Kim et al., 2000). It is important to study an autologous regulatory region in olive flounder for basic research and biotechnological applications such as transgenesis (Beardmore, 1997; Higashijima et al., 1997).
In this study, we isolated and characterized the regulatory region of the ACTC1 gene of olive flounder P. olivaceus (PoACTC1). Pressure-controlled microinjection method was optimized for the delivery of foreign DNA into fish embryo. Its transcriptional activity was shown to direct expression of red fluorescent protein (RFP) in zebrafish and flounder embryos. This study is the first report of autologous promoter-driven transgene expression in the marine fish P. olivaceus.
MATERIALS AND METHODS
Fish were maintained at the Biotechnology Research Division of the National Institute of Fisheries Science (NIFS) in Busan, Korea. Adults (2 years old) were maintained in 1-ton fiber-reinforced plastic (FRP) tanks at an approximate density of 10 fish per tank, with a flow-through system supplied with filtered seawater. Fish were fed a commercial fish diet (crude protein, 52%; crude fat, 11%) three times per day. Fertilized eggs were stocked in a 100-L tank with a flow-through system supplied with filtered seawater along with ultraviolet (UV) light treatment; 98% of the eggs hatched 3 days later. The feeding program was modified from Sakakura (2006). Hatchlings were fed enriched L-type rotifers (Brachionus plicatilis complex) supplemented with chlorella twice a day starting from 3 days post hatching (dph). The temperature of the rearing tanks was maintained at 20℃. Animal experiments were conducted in accordance with the Animal Protection Act of the Ministry of Agriculture, Food and Rural Affairs, Korea, and were approved by the NIFS (2017-NIFS-IACUC-09). For microinjection into zebrafish, fish were maintained at 28.5℃ with a 14-h light/10-h dark cycle, as described by Westerfield (1995).
Expressed sequence tag (EST) clones were isolated from a P. olivaceus cDNA library using a plasmid purification kit (Intron, Seongnam, Korea), and sequenced using universal T3 reverse primer (Promega, Madison, WI, USA) on an ABI3730xl automatic sequencer (Applied Biosystems, Foster City, CA, USA). The EST clone 1before-3-2a-K12showed significant sequence homology to known alpha-actin sequences. The cDNA sequence was annotated in GenBank using BLASTX (http://www.ncbi.nlm.nih.gov/BLAST/). The PoACTC1genomic sequence was predicted by AUGUSTUS, a web server for gene prediction in eukaryotes (Stanke & Morgenstern, 2006), using the genome sequences of P. olivaceus (Kim et al., unpublished data).The potential transcription factor binding sites in the PoACTC1 gene using the PROMO program version 3.0 (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3). The parameters such as “species”, “factors” and “matrix” were set as “All”, and “maximum matrix dissimilarity rate” was 15.
To create the RFP reporter plasmid for the regulatory region of PoACTC1, DNA fragments ranging from positions -2,126 to +751 in Po-actc1 were generated by PCR using Vent DNA polymerase (New England BioLabs, Ipswich, MA, USA) and then inserted into the SalIand AgeI restriction sites upstream of the DsRed2 gene in pDsRed2-1 (Clontech Laboratories, Mountain View, CA, USA). The primer sequences were as follows: Po-ACTC1-2126 FS, 5′-GCC GTC GAC GTT CTT TAA AGG GCT TCT ACT CTG TTT-3′; and Po-ACTC1+751-RA, 5′-AAT ACC GGT GGT GTC AGC TCT GCA ACA CAC ACG GAG-3′. The construct was confirmed by sequencing.
The fluorescent protein reporter construct of the regulatory region of PoACTC1 (pPoACTC1-2877bp) was digested with the restriction endonuclease SalI (New England BioLabs). The purified DNA solution (25 ng/uL) was injected into one-cell-stage zebrafish embryos (n=100) using an air pressure microinjector (PicoPump820, World Precision Instruments, Sarasota, FL, USA), as described previously (Kong et al., 2013). For the microinjection into olive flounder, the method of preparing microinjection needle modified to that described in Goto et al. (2015). Glass microinjection needles were produced by pulling of a borosilicate glass capillary tubes (World Precision Instruments, Inc., Sarasota, FL, USA) using micropipette puller device (Narishige, Tokyo, Japan). End of the tip was melted slightly in a loop of platinum wire to form constriction preventing backflow of cytoplasm and grinded with micropipette grinder (Narishige) to permit smooth penetration into the cytoplasm. The DNA solution (25 ng/uL) was injected into two-cell-stage flounder embryos (n=100) (Kim et al., unpublished data). The estimated average amount of DNA delivered to each embryo was 15-20 pg. Digital images of the embryos were captured using a macro zoom fluorescence microscope (MVX10; Olympus, Tokyo, Japan). The expression rates of red fluorescence were calculated with randomly chosen embryos.
RESULTS AND DISCUSSION
We isolatedPoACTC1cDNA from the olive flounder P. olivaceus by screening a flounder cDNA library (data not shown). The EST clone 1before-3-2a-K12, which carries a 1,336-bp insertion, showed significant sequence homology to known alpha-actin sequences, including a predicted flounder actin mRNA sequence (GenBank no. XM_020111908). The PoACTC1 genomic sequence (4,117 bp) was predicted by the AUGUSTUS gene prediction software and the obtained sequence was confirmed by PCR and sequencing. The sequence contained the PoACTC1gene, covering from 2,126 bp upstream of the putative transcription start site to the fifth exon of PoACTC1 cDNA, which includes the first introns.
The muscle-specific expression of the cardiac alpha-actin gene requires transcription factors such as MyoD1, CArG box binding factor, and Sp1 upstream of the TATA box (Gustafson & Keds, 1989; Sartorelli et al., 1990). We identified potential transcription factor binding sites in the PoACTC1gene using the PROMO program version 3.0. The proximal promoter regionof PoACTC1contains putative regulatory elements such as a TATA box (TATAAATT) at -21 to -14 bp, a MyoD binding site at -155 to -148 bp (ACAGATGT), and two CArG boxes at -78 to -67 bp (CCAAATATGGAG) and -176 to -165 bp (CTCCATAGATGG) from the putative transcription start site (Fig. 1). The pentameric GCTGC sequence associated with the MyoD binding site (Sartotrlli, 1990) was also found in the regulatory region of the PoACTC1 gene. The proximal promoter region of the HCA gene, -117 bp from the start site of transcription, is sufficient to convey muscle-specific expression to heterologous reporters (Minty & Keds, 1986).
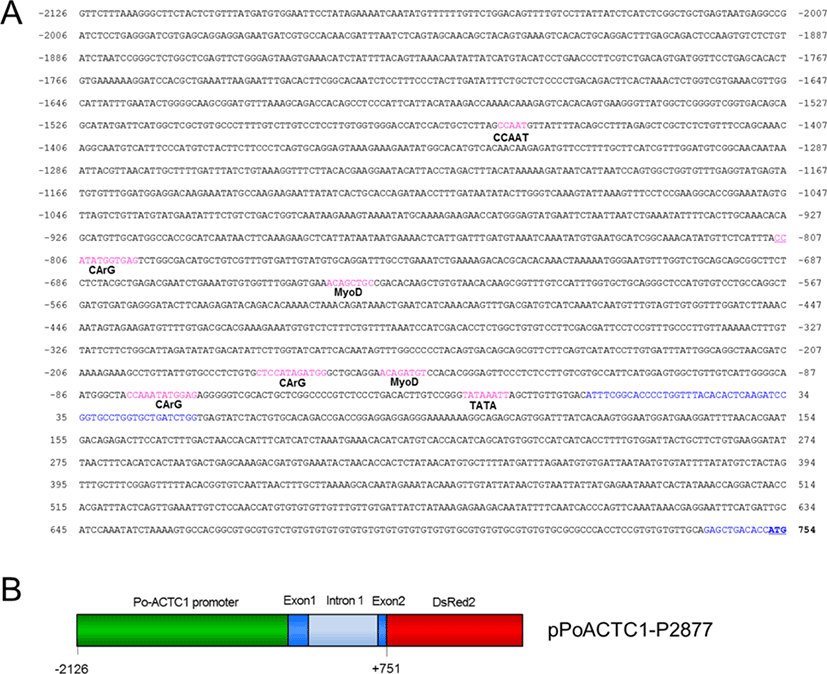
The PoACTC1 promoter also contains various transcription factor binding elements, including an additional CArG-box at -804 to -797 bp, a MyoD binding site at -644 to -637 bp, and a CCAAT box at -1,455 to -1,451 bp in the 5′ upstream promoter region. Several of these elements contribute to the cooperative transcription-activating interaction of the cardiac alpha-actin gene in cellular processes for cell motility, structure, and integrity (Miwa & Kedes, 1987; Bunnell et al., 2011). Further studies may reveal the detailed function and related mechanism of regulatory element (s) in the expression of PoACTC1.
To examine activation of the regulatory region of PoACTC1, we performed a fluorescence microscopy analysis using RFP expression driven by the PoACTC1 regulatory region in zebrafish and olive flounder muscle. In zebrafish, between 9.8% (3 out of 51) and 46.9% (23 out of 49) of the surviving embryos microinjected with pPoACTC1-P2877 exhibited red fluorescence at 2 day post-fertilization (dpf) (Fig. 2A). In olive flounder, pPoACTC1-P2877 also drove RFP expression at 2 dpf, and the signal was still detected at 6 dpf (Fig. 2B). The expression rates of the pPoACTC1-P2877 constructs in olive flounder were between 58.5% (24 out of 41) and 60% (12 out of 20) at 6 dpf. Various techniques have been developed to perform gene delivery of exogenous DNA, including microinjection and electroporation. Microinjection of DNA into fish embryos began from its initial development in fertilized rainbow trout (Oncorhynchus mykiss) in the 1980s (Mclean et al., 1984). Although many studies have reported gene delivery and transgenesis in fish, from model fish such as zebrafish and medaka to aquaculture species such as Nile tilapia and common carp (Tonelli et al., 2017), this is the first report of microinjection and expression of a foreign gene driven by an autologous promoter in embryos of the marine fish olive flounder. We have modified the microinjection protocols to overcome the problem that the hard chorion of its embryo makes microinjection complicated. The success of microinjection and identification of autologous tissue-specific promoter would be influential for basic and applicative studies, which might be helpful to improve the valuable trait of olive flounder.
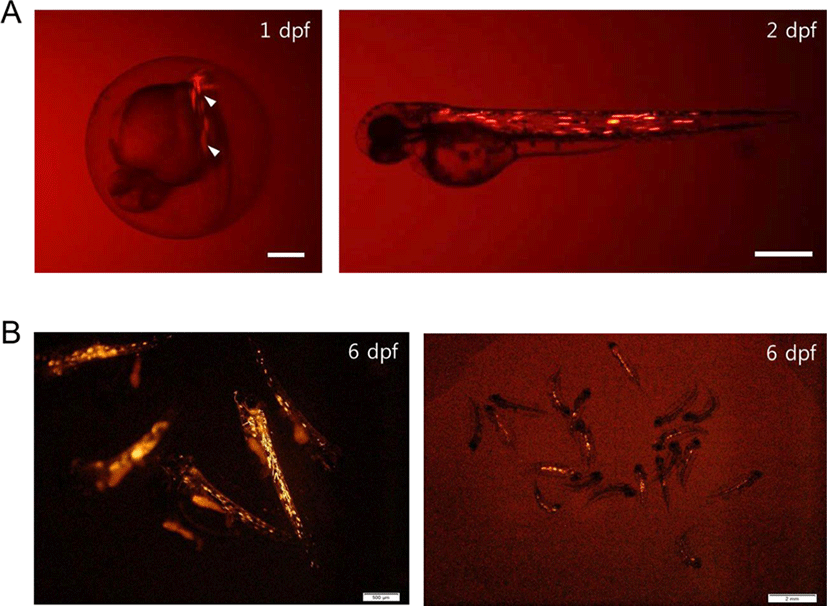
Here, we described the cloning and characterization of the PoACTC1 regulatory region in olive flounder P. olivaceus, a major aquaculture fish in Korea. We demonstrated the activation of the PoACTC1regulatory region in a fluorescence reporter assay using microinjection into zebrafish and olive flounder embryos. These results lay the groundwork for the development of a useful promoter for transgenic technology to develop and improve the traits of this species.
