INTRODUCTION
Penaeid shrimp (Penaeus chinensis), environmentally important aquacultural species, belonging to family Penaeidae including genus Fenneropenaeus, widely distributed on the coast of the Yellow Sea and the southern sea in the Korean peninsula and the Gulf of Pohai in China under the natural ecosystem. Under the natural ecosystem, the shrimps inhabit in the flats consisting of a lot of mud, silt and slime in the coast where the nutrient salts are abundant. Like other shrimps essentially, the rate at which the shrimp grows depends very much on water quality. Largely, there are marked differences of the size, color, shape, disease resistance and mortality in the penaeid shrimp together with the ecological circumstances of habitat such as nurture and rigid period. In spite of its economic and scientific consequence, little information is available on the genetic characteristics among various penaeid shrimp populations in Korea. Thus, the applications of PCR techniques to aquaculture had been to identify genetic distances among a few of fish species and/or invertebrates apart from geographic sites (Partis & Wells, 1996; Callejas & Ochando, 1998; Tassanakajon et al., 1998; Klinbunga et al., 2000; McCormack et al., 2000; Song & Yoon, 2013; Oh & Yoon, 2014; Yoon, 2015). As well, the genetic variation, the species-specific markers and the region-specific markers in finfish have been evaluated by molecular biological methods (Park et al., 2005). Consequent of the rapid increase in seed production, there is a need to understand the genetic traits and composition of this shrimp species in order to evaluate exactly the patent genetic traits induced by seed production operations. This PCR analysis is a quick and reliable method for identifying genetic distances among various life organisms with the advantage that no prior knowledge of the genome under research is needed (Welsh et al., 1991; Partis & Wells, 1996; Mamuris et al., 1999; Nebauer et al., 2000; Yoon & Kim, 2004; Oh & Yoon, 2014; Yoon, 2018). Here, the author undertook clustering analyses in order to elucidate the Euclidean genetic distances and the differences between the fresh and deceased penaeid shrimp populations from Shinan. This author also analyzed the genetic diversity of two shrimp populations in Korea.
MATERIALS AND METHODS
Muscle tissues were collected separately from two fresh shrimp population (FSP) and deceased shrimp population (DSP) shrimp populations, respectively. This shrimp muscle was collected in sterile tubes, placed on ice immediately, and stored at −80°C until needed. DNA samples extracted from a total of 22 individuals. DNA extraction should be performed according to the separation and extraction methods (Song & Yoon, 2013). After washing several washings, samples of muscle tissues were placed into 10 ml test tubes, to which 3 volumes of lysis buffer (155 mM NH4Cl, 10 mM KHCO3, 1 mM EDTA) was added, and the mixture tubes were lightly inverted. The precipitates obtained were diffused with lysis buffer (10 mM Tris-HCl, pH 8.0, 10 mM EDTA, 100 mM NaCl, 0.5% SDS). Samples were added 15 µl proteinase K solutions (10 mg/ml). After incubation, there was added 300 µl of 6 M NaCl and softly pipetted for a few of min. 600 µl of chloroform was added to the mixture and then inverted (no phenol). The DNA pellets were incubation-dried for 2 hrs, held at −80°C until analysis, and then dissolved in the TE buffer (10 mM Tris-HCl, pH 8.0; 1 mM EDTA). Ice-cold 70% ethanol was added, and then the samples were centrifuged at 19,621 g for 5 minutes to extract DNA from the lysates. The concentrations of the extracted genomic DNA samples were estimated based on the optical density at 260 nm by a spectrophotometer (Beckman Coulter, Buckinghamshire, UK). The DNA pellets were then incubation-dried for more than 12 hours, maintained at −80°C until needed and then dissolved in the distilled water.
Amplification products were separated by electrophoresis in 1.4% agarose gels with TBE, using 100 bp DNA ladder (Bioneer Corp., Daejeon, Korea) as DNA molecular weight marker and detected by staining with ethidium bromide. The electrophoresed agarose gels were illuminated by ultraviolet rays, and photographed using a photoman direct copy system (PECA Products, Beloit, WI, USA). The author used the oligonucleotides primers to clarify the genetic distances and variations of P. chinensis individuals. Seven primers (Operon Technologies, Alameda, CA, USA), OPA-03 (5′-AGTCAGCCAC-3′), OPA-07 (5′-GAAACGG GTG-3′), OPA-09 (5′-GGGTAACGCC-3′), OPA-10 (5′-GTGATCGCAG-3′), OPA-18 (5′-AGGTGACCGT-3′), OPA- 19 (5′-CAAACGTCGG-3′), and OPA-20 (5′-GTTG CGATCC-3′) were displayed to generate the unique shared loci to each population and shared loci by the two P. chinensis populations which could be visibly counted. Thus, the author used the oligonucleotides primers to determine the genetic differences and variations of the penaeid shrimp. PCR was performed using programmable DNA Thermal Cycler (MJ Research Inc., Waltham, MA, USA). Similarity matrix including bandsharing values between different individuals in the two penaeid shrimp populations was created allowing formula of Jeffreys & Morton (1987) and Yoke-Kqueen & Radu (2006). The average of within-population similarity was calculated by pairwise comparison between individuals within a population. A hierarchical dendrogram was assembled using similarity matrices to yield a cluster tree, which was supported by the Systat version 10 (SPSS Inc., Chicago, IL, USA). The Systat software was also used to analyze genetic distances, Euclidean distances within and between shrimp populations, means, standard errors, and t-test scores.
RESULTS AND DISCUSSION
The number of total loci, average loci per lane and specific loci was investigated by analytical technique using 7 oligonucleotides primers from two penaeid shrimp individuals from FSP and DSP population, respectively, as showed in Table 1. Here, the complexity of the banding patterns varied dramatically between the primers from the two sites. The size of DNA fragments also varied roughly, from 100 to 1,800 bp. In this study, 233 loci were identified in the FSP shrimp population and 162 in the DSP shrimp population: 33 specific loci (14.2%) in the FSP shrimp population and 42 (25.9%) in the DSP population. These results exhibit that the primer detected a large number of specific fragments, suggesting that the genetic variation in the DSP is higher than in the FSP shrimp population. Especially, OPA-10 oligonucleotide primer gave DNA profiles with more fragments than the other six primers in the FSP shrimp population. The oligonucleotides primer OPA-03 generated 4.1 average loci per lane in FSP shrimp population. Also, the oligonucleotide primer OPA-18 produced 18 specific loci in the DSP shrimp population. A total of 42 (an average of 6.0 per primer) were observed in the DSP shrimp population, whereas 33 specific loci (an average of 4.7 per primer) in the FSP shrimp population. Especially, the specific markers unique to species, breed and population have been used for individual or species identification, genetic origin of species, population genetics, screening of traits like disease resistance and for breed improvement in breeding programs (Ramesha et al., 2002). Generally, the specific oligonucleotides primer could be used for detecting genetic similarity/diversity/poly-morphisms among various organisms (Welsh et al., 1991; McCormack et al., 2000; Nebauer et al., 2000; Zhou et al., 2000; Ramesha et al., 2002; Yoon & Kim, 2003a; Kim et al., 2004; Song & Yoon, 2013). A total of 66 (an average of 9.4 per primer) were observed in DSP shrimp population, whereas 55 unique loci to each population (an average of 7.9 per primer) in the FSP shrimp population, as summarized in Table 2. The oligonucleotides primer OPA-09 generated 22 unique loci to each population in the DSP shrimp population. Interestingly, the other primers had not distinguished the shared loci by the two populations excluding OPA-09. Marvelously, the primer OPA-09 detected 22 shared loci by the two populations, major and/or minor fragments, which were matching in all samples. Several investigators researched the sizes of DNA fragments in the PCR frameworks of black tiger shrimp (Penaeus monodon) (Tassanakajon et al., 1998), Eastern Pacific abalone (genus Haliotis) (Muchmore et al., 1998), the brittle star (Amphiura filiformis) (McCormack et al., 2000), shrimp populations (Yoon & Kim, 2003b), and Pollicipes mitella population (Song & Yoon, 2013).
Parentheses are the total number of products generated in fresh and deceased shrimp population, respectively.
FSP, fresh shrimp population; DSP, deceased shrimp population.
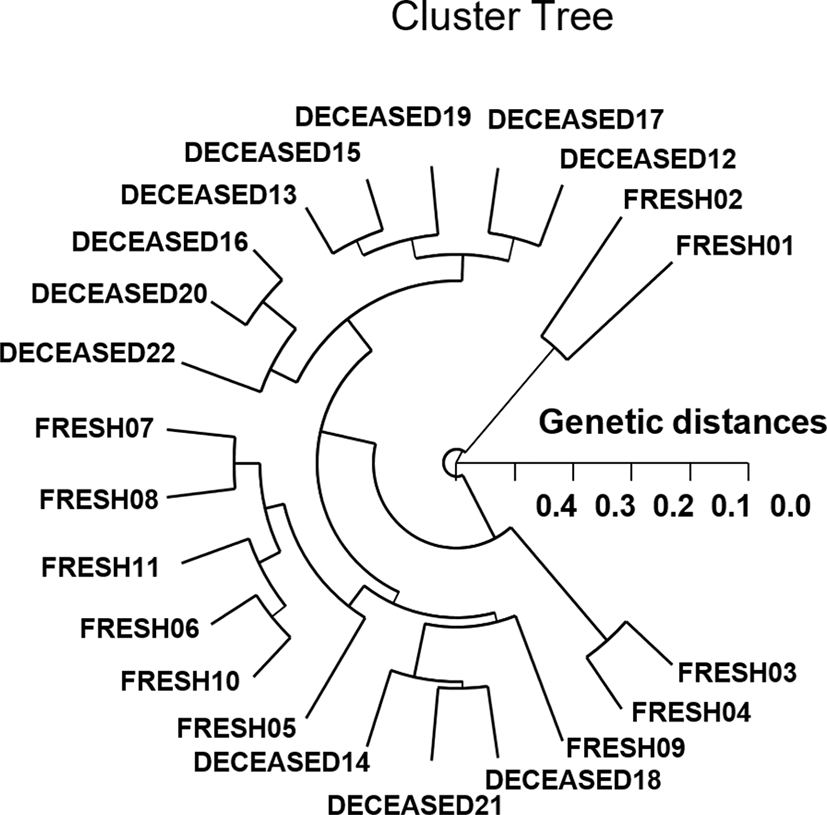
As regards average bandsharing value (BS) results, individuals from FSP shrimp population (0.654) displayed higher bandsharing values than did individuals from DSP population (0.592) (p<0.05), as demonstrated in Table 3. In the present study, the Hierarchical dendrogram extended by the seven oligonucleotides primers indicates three genetic clusters: cluster 1 (FRESH 01, 02, and DECEASED 12, 13, 15, 16, 17, 19, 20, 22) and cluster 2 (FRESH 03, 04, 05, 06, 07, 08, 09, 10, 11, and DECEASED 14, 18, 21), as shown in Fig. 1. Among the twenty-two shrimp, the shortest genetic distance that exposed significant molecular differences was between individuals 20 and 16 from the DSP shrimp population (genetic distance=0.071), while the longest genetic distance among the twenty-two individuals that established significant molecular differences was between individuals FRESH no. 02 and FRESH no. 04 (genetic distance=0.477). In due course, PCR analysis has revealed the significant genetic distance among two penaeid shrimp populations by PCR-based approach. In invertebrates, cluster analysis of the pairwise population matrix, generated from genetic data, showed that geographically close populations be inclined to cluster together in the blacklip abalone (Huang et al., 2000). PCR fragments exposed in this study could be valuable as a DNA marker of the two shrimp populations to discriminate. The potential of oligonucleotides amplified polymorphic DNAs to discover diagnostic markers for line, breed, species, genus, and population identification in shellfish (Huang et al., 2000; Klinbunga et al., 2000; McCormack et al., 2000; Kim et al., 2004; Park et al., 2005; Song & Yoon, 2013; Oh & Yoon, 2014) has also been well established. High levels of a significant genetic distance between two penaeid shrimp populations showed this PCR approach is one of the most suitable tools for individuals and/or populations biological DNA studies (Callejas & Ochando, 1998; Tassanakajon et al., 1998; Mamuris et al., 1999; Nebauer et al., 2000; Yoon & Kim, 2003a; Yoon & Kim, 2004; Yoon, 2018).
| Population | FSP | DSP |
|---|---|---|
| FSP | 0.654±0.011a | 0.574±0.009b |
| DSP | − | 0.592±0.021ab |
Each value is a result of three different experiments.
FSP, fresh shrimp population; DSP, deceased shrimp population.
