INTRODUCTION
Spermatozoa are derived from the spermatogonia through the mitotic proliferation of spermatogonial stem cells (SSCs), meiosis, and spermiogenesis. The ancestral cells of SSCs are gonocyte and primordial germ cell (PGC) in the testis cord and the sex cord, respectively. The germline stem cell is one of the gonadal stem cells, the PGC, and is derived from the extraembryonic mesoderm at the caudal end of the primitive streak. It is first specified around embryonic day 6.25 (Hill et al., 2018) and can be detected in early 8-day embryos by testing alkaline phosphatase activity (Chiquoine, 1954). In males, these cells differentiate into gonocytes between E12.5 and E14.5 and remain at the G1/G0 phase in an unsynchronized manner during E13.5–E14.5 (Garcia & Hofmann, 2013). Gonocytes develop into SSCs approximately 6 days after birth in rodents. Proliferation of the SSCs and spermiogenesis is then observed from the pubertal stage.
During spermatogenesis and further development, genome-wide DNA methylation and imprinting occur at the PGC for gonocyte transition (Hill et al., 2018; Ueda et al., 2000). There is near complete loss of combined 5-methylcytosine and 5-hydroxymethylcytosine (5 mC/5 hmC) between E10.5 and E11.5, with limited DNA demethylation between E11.5 and E12.5 (Hill et al., 2018). Selective usage of inherited information is a key component of development and the preparation of the gamete. In Drosophila melanogaster, the male germline-controlling genes are found in transcriptionally silent chromatin and Polycomb-group proteins (PcGs) are required to maintain this silent state and to regulate selective usage of these genes (Gan et al., 2010). PRC2 (Polycomb repressive complex 2), one of the two classes of PcGs, is a key regulator of proliferation and differentiation via gene silencing (Orlando, 2003; Mu et al., 2014). The other component of this group of proteins, PRC1, is involved in long-range intra- and inter-chromosomal interactions between Polycomb target loci. It functions as a master regulator of genome architecture, regulating stem cell self-renewal (Schoenfelder et al., 2015). In mammals, CpG islands, long noncoding RNAs, and Polycomb response elements (PREs) are targets of PcGs (Simon & Kingston, 2013). The functions of PcGs at these targets depend on the multi-protein complexes that selectively occupy the chromatin sites.
Cell type-specific establishment and maintenance of cell identity are key steps in development. Germ cells are generally considered to be totipotent (Seydoux & Braun, 2006). Epigenetic changes during the gonocyte transition recapitulate complete gametogenesis (Hill et al., 2018). Perturbation of the epigenome during transition of PGCs to gonocytes might ultimately impede spermatogenesis. Spermatogenesis in mammals requires programmed expression based on circuits of transcription factors (Wang et al., 2017). These circuits induce chromatin structural changes and epigenetic reprograming. PcG complexes are key regulators of epigenetic reprogramming and interact with various epigenetic modulators, including transcription factors.
Although chromatin structural modification by Polycomb is essential for gene silencing in mammals, and epigenetic changes are critical in spermatogenesis, a comprehensive understanding of the transcriptional regulation of spermatogenesis is lacking. A non-canonical polycomb repressor ncPRC1.6 include Yin YY1 and RING1 as a core member, and plays in control of germ-cell specific genes. Recently, we evaluated the distribution of YY1 and CP2c (alpha-globin transcription factor CP2, TFCP2; α-CP2; LSF; LBP-1c) in male germ cells (Kim et al., 2016). In the present article, we discuss the possible roles of YY1 and CP2c in spermatogenesis and potency regulation by reviewing the relevant literature.
SPERMATOGENESIS REGULATORS
PcG proteins are part of the multi-protein complexes PRC1, 2/3 and pliohomeotic repressive complex. PRC1 forms a diverse number of complexes. Canonical CBX-PRC1 comprises CBX (polycomb), PCGF (polycomb group factor; polycomb group RING finger protein 4, PCGF4; RING finger protein 51, RNF51; B cell-specific moloney murine leukemia virus integration site 1, BMI-1), HPH (human polyhomeotic homolog; PHC), and the E3 ubiquitin protein ligase protein (RING1A/B). These proteins have two to six paralogs, leading to diverse CBX-PRC1 compositions (Klauke et al., 2013; Connelly & Dykhuizen, 2017). Non-PcG interactors (kinases, general transcription factors, etc.) also interact with this canonical complex. This PRC1 diversity may regulate specific sets of target genes, thereby activating diverse targets due to different binding affinities (Gil & O’Loghlen, 2014). PRC2 has following components, SUZ12 (suppressor of zeste 12), EED (embryonic ectoderm development), EZH1/2 (enhancer of zeste 1/2), JARID2 (Jumonji, ARID domain-containing protein 2), and Rbbp7/4 (Rb binding protein 7/4) (Orlando, 2003; Simon &, Kingston, 2009). This complex interacts with long noncoding RNA (Simon & Kingston, 2013). It has been hypothesized that specific PcG proteins are epistatic in the control of other PcG members (Orlando, 2003). In some cases, PRC2 interacts with the canonical PRC1 to silence genes (Di Croce & Helin, 2013). PhoRC consists of Pho and DSFMBT subunits (Gautam et al., 2020). PcG modifies chromatin structure by promoting histone methylation or mono-ubiquitination, preserves epigenetic memory, and controls transcriptional programs (Simon & Kingston, 2009; Surface et al., 2010). Mutations of PcG genes cause of dysfunction in differentiation of germ cells, of lost the gene expression for germ-cell-specific genes (Yokobayashi et al., 2013; Maezawa et al., 2017), and of altering reprogramming processed in oocyte and stem cells (Li et al., 2017; Du et al., 2020). Maintaining the undifferentiated phenotype in embryonic stem cells (ESCs) is generally ensured by PRC-mediated repression of developmental genes (Surface et al., 2010). However, PcG also permits specification through activation of specific gene sets (Abed & Jones, 2012; Blackledge et al., 2015; Lee et al., 2018).
YY1 is a ubiquitously distributed transcription factor belonging to the GLI-Kruppel class of zinc-finger proteins. YY1 is involved in repressing and activating a diverse number of promoters, and it interacts with positive and negative transcriptional regulators, either dependent or independent of its DNA binding affinity (Deng et al., 2010; Infante et al., 2015). YY1 may direct histone acetyltransferases and histone deacetylases to a promoter in order to activate or repress the promoter (Coull et al., 2000), thus implicating histone modification in the function of YY1. YY1 also functions as a component of PcG complexes, mediating their binding ability to the PRE (Satijn et al., 2001). YY1 regulates the expression of many genes, including c-Myc, c-Fos, IFN-, CREB, and Sp1 (Wang & Goff, 2015). One of the mechanisms used to suppress YY1 is to reduce its effects on target genes, like Peg3, by reduced expression 1 (REX1), an epigenetic regulator of genomic imprinting during oogenesis (Kim et al., 2009; He et al., 2017). Rex1 encodes a C2H2 zinc-finger protein and it has high homology with YY1. It is only expressed during spermatogenesis and early embryogenesis and in a very limited area of the placenta. Interestingly, YY1 and REX1 have opposite roles in the epigenetic imprinting of Peg3, through the differential recruitment of PRCs. YY1 induces the methylation of the differentially methylated region (DMR) of Peg3 during oogenesis, whereas REX1 demethylates it (Kim et al., 2011). However, YY1 is mainly involved in controlling the transcriptional levels, not the DNA methylation, of paternally inherited Peg3 (He et al., 2017). In addition, it was recently found that YY1 directly interacts with another transcription factor, CP2c (Coull et al, 2000; He & Margolis, 2002), and these mutually suppress expression of their target genes (Kang et al., 2005; Kim et al., 2016).
The CP2c gene encodes a transcription factor that binds to the alpha-globin promoter and activates transcription of the α-globin gene (Kim et al., 1988, 1990). The CP2c protein regulates erythroid gene expression, plays a role in the transcriptional switch of globin gene promoters (Chae & Kim, 2003), and activates many other cellular and viral gene promoters (Veljkovic & Hansen, 2004). CP2c belongs to the CP2 family of proteins, with six isoforms (LBP-1a, −1b, −1c, −1d, −9 and LBP-32) in humans and four (CP2a, CP2b, CP2c, and CRTR-1) in mice (Kang et al., 2005). CP2c binds directly to the target DNA sequences, CNRG-N6-CNR (G/C), in the regulatory regions of genes, including the α - and β -globin, GATA-1, and EKLF genes (Kang et al., 2010). CP2c is ubiquitously expressed, but functions as a homotetramer (Murata et al., 1998) or as heteromeric combinations, such as the heterohexameric complex of CP2c, CP2b, and PIAS1 (Kang et al., 2010). CP2c complexes can act as transcriptional activators or inhibitors in various cellular processes and in malignant diseases, such as cancer, Alzheimer’s disease, and hemoglobinopathies (Veljkovic & Hansen, 2004; Santhekadur et al., 2012), depending on the other components of the complex, including YY1, RING1, RNF2, HDAC1/2, SIN3A, Fe65, NF-E4, TTRAP (DPF2), GATA1, GATAD2A, PIAS1, SUMO1, and UBP9 (Kang et al., 2010; Kim et al., 2017). In addition, these CP2c complexes recruit chromatin-remodeling complexes (CRCs) via direct interaction with their components, p66 α (GATAD2A) in NuRD CRC (Kim et al., 2018) and BRD7 in BAF (unpublished), conferring CP2c with diverse regulatory functions in a context-dependent manner.
EXPRESSION OF YY1 AND CP2C DURING GONADAL FORMATION
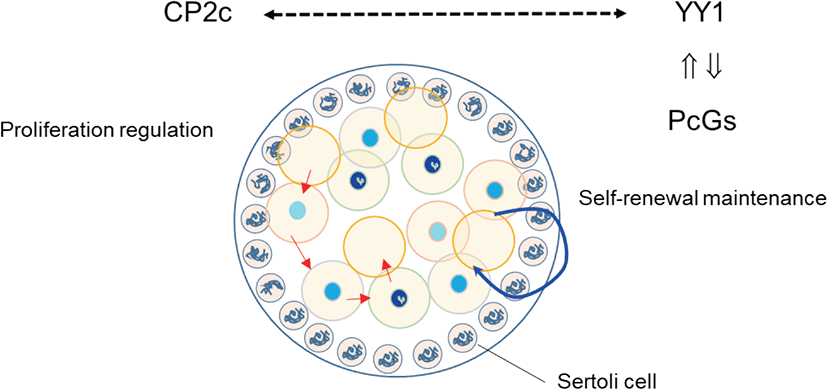
We previously evaluated the localization of YY1 and CP2c during male gonadogenesis (Kim et al., 2016). YY1 and CP2c are localized heterogeneously to gonocytes of the testicular cord at the E14.5 gonadal ridge. YY1 is mainly localized in gonocytes that have densely stained heterochromatin particles in the nucleus, whereas CP2c localizes in gonocytes with weak intensity. It is stained mainly in cytoplasm of gonocytes. Based on neighbor section immunohistochemistry, they do not colocalize in the nucleus of the mitotically inactive gonocytes (Fig. 1). Mesenchymal cells surrounding the testicular cords have nuclei that stain strongly for both YY1 and CP2c, with faint cytoplasmic staining. Localization of YY1 and CP2c in gonadal somatic cells is heterogeneous, as in gonocytes. YY1 can induce glial cell line-derived neurotrophic factor (GDNF)-induced survival and self-renewal of mouse SSCs (Wu et al., 2010). PRCs can couple chromatin-PcG complexes and the DNA of target genes via YY1 (Cavalli & Paro, 1998), and CP2c works antagonistically to YY1 (Kang et al., 2005). In a 14.5 dpc embryo, gonocytes remain at the mitotic stage (Culty, 2009). Together, these data suggest that the YY1-PRC interaction, independent of CP2c, is involved in the generation of new imprinting or genome-wide methylation of gonocytes and maintenance of heterochromatin integrity in gonocytes. In addition, it is suggested that the shuttling of CP2c may involve in mitosis with YY1. We intend to explore if YY1 and CP2c are involved in self-renewal, regulation of mitotic proliferation, and stem cells competency by interaction with PcGs in future studies.
YY1 AND CP2C FUNCTION IN PLURIPOTENT STEM AND GERM CELLS
PGCs can change their fate and be reprogrammed into pluripotency in culture. Reprogramed germ cells become pluripotent stem cells like ESCs. However, adult SSCs resist reprogramming induction by Yamanaka’s factors (Oct3/4, Sox2, Klf4, and c-Myc), and it has been suggested that there is a specific mechanism that prevents reprograming by these factors (Corbineau et al., 2017).
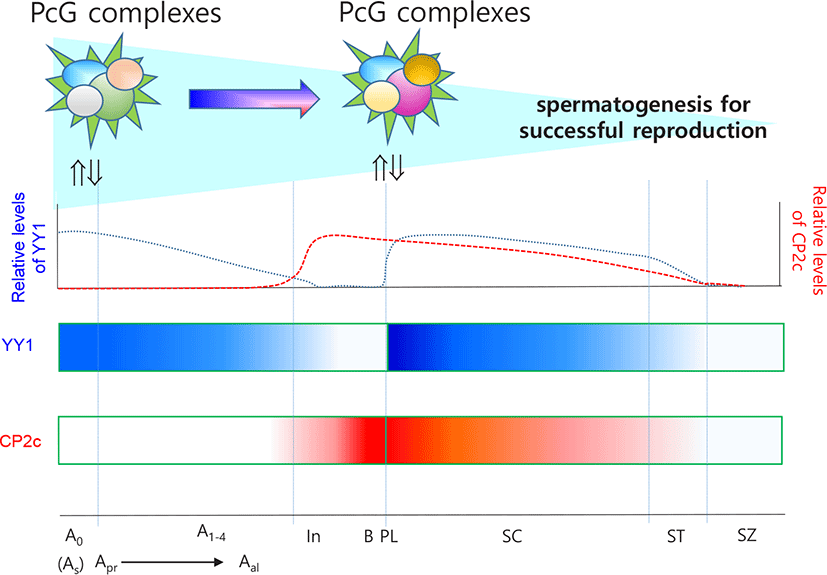
SSCs are defined as GFRα1 (GDNF family receptor alpha 1)-positive undifferentiated type A (Aundiff, A0) cells, which exist as single cells (Asingle or As). Pairs of spermatogonia (Aparied or Apr) connected by a cytoplasmic bridge or chains of 4- to 16-cell spermatogonia (Aaligned or Aal) are undifferentiated with commitment for differentiation (Barnhart et al., 1988; Nakagawa et al., 2010). FOXC2 (Forkhead box C2) is expressed predominantly in As and Aparied spermatogonia, and knock-down of Foxc2 decreases SSC colonization (Wei et al., 2018). Differentiation begins when Aal cells transition to c-KIT-positive spermatogonia (de Rooij & Grootegoed, 1998). A1 spermatogonia (committed SSCs) subsequently undergo five additional cell cycles (A2, A3, A4, Intermediate (In), and type B spermatogonia). GFR α 1 expression may be crucial for type A spermatogonia because Gfra1 knockdown leads to differentiation of these spermatogonia (He et al., 2008). The final type B spermatogonia differentiate into spermatocytes that undergo meiosis. A type spermatogonia can be distinguished as Adark (Ad), Apale (Ap), and At, especially in primates. Ad spermatogonia are mitotically quiescent in the post-pubertal testis as a stem cell reserve. Ap spermatogonia are mitotically active and classically considered to be renewing stem cells.
Such characteristics of spermatogenesis follow the principles of development and allow the induction of stem cells into a specific cell lineage or dedifferentiation. Although current knowledge cannot explain why SSCs do not easily transform into ESCs, investigating the interactions of transcription factors and chromatin modulators may lead to an understanding of the unidirectional process in germ cells. Doublesex and mad-3-related transcription factor 1 (DMRT1) supports SSC maintenance, allowing SSCs to remain in the stem cell pool under normal conditions and enabling progenitor cells to help restore the stem cell pool after germ cell depletion (Zhang et al., 2016). However, expression of genes that encode proteins that suppress methylation of histones and DNA is a primary event in totipotent embryos, and large-scale selective silencing is observed (Orlando, 2003). During development, histone tails undergo acetylation, sumoylation, phosphorylation, ubiquitination, and methylation (Gil & O’Loghlen, 2014).
Among the regulators of chromatin dynamics is PcG. Genes targeted by PcG and the trithorax group (trxG) prevent non-specific transcription and maintain transcription patterns in developing and specified cells. PcG proteins maintain the chromatin in an inactive condition, creating a heterochromatin-like environment, by methylation mediated by methyltransferase-containing PRC1 and 2 of H3-K27 (Orlando, 2003; Simon & Kingston, 2009). PRC2 and H3K27me3 inactivate developmental regulators (Lee et al., 2006). Indeed, PcGs play critical roles in determining the potency of ESCs and adult stem cells and are involved in the differentiation of somatic cells (Ezhkova et al., 2009). PRC1 and 2 work as major regulators of genome architecture changes required for stem cell self-renewal via a cell-autonomous mechanism (Simon & Kingston, 2013; Schoenfelder et al., 2015; Chiacchiera et al., 2016) by acting as major mediators of gene silencing and heterochromatin formation (Margueron & Reinberg, 2011). The expression of differentiation genes is repressed by PRC2 in adult stem cells (Ezhkova et al., 2009). It is also noted, however, that PcG proteins can coexist and include activators to change cell fate (Orlando, 2003). In mouse ESCs, the Kdm2B-PRC1 complex is known to act as a transcriptional activator and promote self-renewal by targeting early lineage-specific genes, depending on the recruitment and maintenance of PRC1 (Barrero et al., 2013).
Few papers have investigated the possible role of YY1 in stemness maintenance (Pan et al., 2012; Vella et al., 2012), where YY1-related PRCs are considered key players in stemness (Perekatt et al., 2014). Because one of the presumed candidates of PRC2-targeting factors is YY1, the interactions between YY1 and PRC1 and 2 inhibit chromatin remodeling for transcriptional activation (Onder et al., 2012). In bone marrow cells, overexpression of YY1 inhibits the progression from hematopoietic stem/progenitor cells to pro-B, pre-B, immature B, and re-circulating B cell stages, which allows the enrichment of long-term hematopoietic stem cells (Pan et al., 2012). YY1 is also an essential transcription factor for long-term self-renewal of intestinal stem cells that reside in intestinal crypts; these cells give rise to all other intestinal epithelial linages (Clevers, 2013), and lack of YY1 in these stem cells results in stem cell depletion (Perekatt et al., 2014). Thus, YY1 plays a role in cell proliferation and differentiation as a stemness factor (Perekatt et al., 2014). However, as a Myc-related transcription factor, YY1 can also enhance the expression of highly expressed genes in ESCs (Vella et al., 2012). YY1 also has RNA helicase and genome-wide PcG-independent activities by stable association with all components of the INO80 CRC in mouse ESCs (Vella et al., 2012). Furthermore, inhibition of the core components of PRC1 and 2 (such as the H3K27 methyltransferase EZH2) reduces reprogramming efficiency for induced pluripotent stem cells, which involves global epigenetic remodeling, and coordinates with chromatin modifying proteins, whereas YY1 enhances reprogramming efficiency (Onder et al., 2012), suggesting another PRC1 and 2-independent function of YY1. These observations suggest that the role of YY1 in stemness is complicated, and that further research is warranted.
Mu et al. (2014) and Orlando (2003) suggested that, in male germ cells, PRC2 regulates proliferation and differentiation through gene silencing. Indeed, PRC2 is essential for maintenance of SSCs (Mu et al., 2014), while PRC1 and Tet1 enable the activation of a critical set of germline reprogramming responsive (GRR) genes involved in gamete generation and meiosis (Hill et al., 2018). Because YY1 can be recruited by CP2c, and YY1 and CP2c have antagonistic effects on transcription (Coull et al., 2000; Kang et al., 2005), we previously evaluated the expression profiles of YY1 and CP2c in adult seminiferous tubules (Kim et al., 2016) based on the EGMR classification of spermatogenesis (Barnhart et al., 1988). YY1 localized at the nucleus of SSCs in all stages but only localized in G and M stages and faintly in R stage in spermatogonia. In contrast, CP2c was not detected in any stage of SSCs but was present in spermatogonia and spermatocytes at all stages. Thus, we suggest that YY1 is essential for the stemness of SSCs via its interaction with PRC2, as for other embryonic and adult stem cells.
SPECIFIC EXPRESSION OF YY1 AND CP2C DURING SPERMATOGENESIS
For a PGC to become a gonocyte, there has to be loss of 5-methyl-cytosine (5 mC) at a genome-wide level (Hill et al., 2018). Such dramatic epigenetic reprogramming by TET-1-mediated 5 mC oxidation occurs in E10.5-E11.5 in mice and prevents the expression of GRR genes. Germline reprogramming is activated through TET-1-mediated demethylation and PRC1 loss in PGC (Hill et al., 2018). After birth, gonocytes resume mitosis and develop into SSCs or A0 spermatogonia (Culty, 2009).
PRC2-mediated gene silencing involves methylation of H3K27 by the RING protein of PRC1 (Di Croce & Helin, 2013). Polycomb can bind directly to a group of spermatogenesis-involved germline genes, such as Sall4, in SSCs (Maezawa et al., 2017). RING1B (RNF2), a component of PRC1, is expressed abundantly in undifferentiated and differentiation-committed spermatogonia. PRC1 is required for the maintenance of undifferentiated spermatogonia, and PRC1 loss of function results in germ cell depletion prior to meiosis due to defective RING1B-dependent activation of germline genes (Maezawa et al., 2017). It is well characterized that RING1 and YY1 binding protein (RYBP) is the germ-cell-specific epigenetic regulatory complex. RYBP is localized in spermatogonia (Bajusz et al., 2019). Furthermore, YY1 expression is strictly restricted to the nucleus of SSCs and germ cells derived from it (Kim et al., 2016). However, CP2c, whose transcriptional activity is inhibited by RING1B binding (Lee et al., 2008), is not expressed in SSCs, suggesting that YY1 is a key factor for maintaining the viability of spermatogonia and the stemness of SSCs as one of the elements of PRC1 in SSCs.
Spermatocytes undergo meiosis I and II. After S phase, the committed spermatogonia become primary spermatocytes and undergo meiosis. For complete meiosis, meiotic recombination begins with programmed double-strand breaks and homologous recombination (Cao et al., 1990). MYBL1 transcription factor functions as a master regulator of male meiotic processes (Bolcun-Filas et al., 2011). MYBL1 appears in the pachytene stage and activates the expression of diverse genes including those involved in nucleotide binding, transcriptional regulation, and the cell cycle. During spermatogenesis, level of H3K27me3 decreases (Sasaki & Matsui, 2008), as do global H3K9me3 and HP1- γ levels in spermatocytes (Wu et al., 2009) in a similar manner in Drosophila, where the combined action of aly- and can-class gene products results in removal of H3K27me3 and PcG and recruitment of RNA pol II to target gene promoters during spermatogenesis (Chen et al., 2011; Laktionov et al., 2014). However, γ H2AX+ meiotic cells of PRC1c KO mice have no XY bodies or histone variant H1T and cannot progress to the mid-pachytene stage, suggesting that PRC1 is also required for meiosis (Maezawa et al., 2017). Accordingly, YY1 localizes to the heterochromatin of spermatocytes and participates in maintenance of stability and homologous recombination-mediated DNA repair (Wu et al., 2009). In germ cell-specific YY1 conditional knockout mice, pachytene cell loss is observed with double-strand break accumulation, and YY1-deficient spermatocytes have decreased heterochromatin and increased double-strand break signals at the leptotene/zygotene stages (Wu et al., 2009). In contrast, YY1 is highly expressed in pachytene, diplotene, and diakinesis stages but not in the leptotene stage and only faintly in the zygotene stage (Kim et al., 2016). YY1 knockdown does not cause any major changes in the DMRs of Peg3 and Xist, the locations of YY1 binding sites(Kim et al., 2009, 2011). These data suggest that YY1, as a partner of PRCs, participates in homologous recombination-mediated DNA repair during meiosis I but does not play a role in the maintenance of methylation status of DMRs. In addition, Cyclin A1 (Ccna1 gene product) present in pachytene and diplotene spermatocytes during spermatogenesis functions as a male germ cell-specific cell cycle regulator. Consistent with the pattern of Ccna1 expression, H3K27me3 enrichment in the promoter takes part in pachytene spermatocytes (Panigrahi et al., 2012). The loss of aggregated PRC1 components can lead directly to chromatin decompaction, and altered structure or the loss of Polycomb interaction leads to a return to a basal or inactive chromatin conformation, followed by further decompaction to the active state as a result of coincident gene depression (Williamson et al., 2016). The overall data support the importance of YY1 as a partner of PRCs in spermatogenesis.
CP2c, in connection with YY1 expression and cellular distribution, is critical for the commitment of spermatogonia and progression of spermatogonia to spermatocytes (Kim et al., 2016). Because spermatocytes undergo meiosis, CP2c and YY1 are expected to have different roles in these cells from those in spermatogonia. Interaction between YY1 and CP2c might be important for expression of spermatogonia-specific genes for meiotic progression. Indeed, rhythmical changes in the localization patterns of YY1 and CP2c in germ cells are meiosis stage specific and indicate general roles for YY1 and CP2c in spermatogenesis (Kim et al., 2016). YY1 is localized in the nucleus and/or cytoplasm of spermatocytes, whereas CP2c is localized in the nucleus, where YY1 and CP2c are colocalized, and YY1 localizes in the cytoplasm of spermatocytes and spermatids (Kim et al., 2016). Transcription factors translocate into the nucleus in a similar manner to that reported during erythroid differentiation and interact with YY1 and CP2c to suppress YY1 activity by protein-protein interactions (Coull et al., 2000; Kang et al., 2005), affecting spermatogenesis-specific gene expression in spermatocytes and spermatids. Based on the available evidence, we suggest that YY1 and CP2c function as key transcription activators to maintain meiotic progression concomitant with other regulators (Fig. 2). This evaluation expands our understanding of the molecular mechanisms of spermatogonia formation as well as spermatogenesis in general.
CONCLUSIONS
The unidirectional progression of male germ cell formation, including stem niche, stemness, and differentiation into meiotic germ cells, is a unique phenomenon in mammalian development. However, the detailed molecular mechanisms underlying male germ cell formation have yet to be elucidated. In addition, the induced dedifferentiation of SSCs using induced pluripotent stem cell strategies has thus far been mostly unsuccessful. Current evaluation of stage-dependent testicular YY1 and CP2c expression during gonadogenesis and the spermatogenic cycle suggests that YY1 is essential for stemness, as a partner of PRCs, whereas CP2c is critical for the commitment of spermatogonia and the progression of spermatogonia to spermatids due to its antagonistic action to that of YY1. The relationship between YY1 and CP2c in spermatogenesis is just starting to be unraveled and not yet fully understood. It is likely that interactions between YY1, CP2c, PRCs, and CRCs fine tune gene expression. CRCs (SWI/SNF, ISWI, CHD, and INO80) evict, slide, insert, or replace nucleosomes, which represent an intrinsic barrier to DNA access and play a role in providing access to the genome by transcription-factors and DNA replication and repair proteins (Clapier & Cairns, 2009). Interplay among CRCs recruited to chromatin via specific interactions with transcription factors, is likely crucial for fine tuning of gene regulation. However, although the four mammalian remodeler families are frequently mutated in cancer (Kadoch & Crabtree, 2015), it remains largely unclear how they orchestrate the global organization of nucleosomes and regulate transcriptional fine tuning. Further studies are required to determine the molecular mechanisms of interconnectedness among proteins in these families as well as their roles in stemness and fate commitment decisions. We are confident that advances in analytical tools will fallow evaluation of genetic information usage during gonadogenesis and gametogenesis.
