INTRODUCTION
This mollusk (Scapharca subcrenata) is one of the most customary crustacean species in winter season of Korea. This invertebrate species is an cost-effectively warm-water shellfish type, being adjacent to the class Bivalvia, order Arocida, family Arcidae, and relatives of the genus Scapharca spread wide-range in the Buan bay, Shinan bay, Suncheon bay, Yeoja bay, and several foreshore areas of Korea. This invertebrate is also home-grown to the sand or mud zones covering slime from 5 to 10m in deepness. The rough lid guard of this invertebrate kind is blackish mud color, vaulted with gloomy hair, with rose-colored muscle in the internal body. Like other shellfish, the great and granular furrow hollows of this invertebrate are affected deliberately by the marine environments. Most of all, there are distinct alterations in the shell volume, shell color, and muscle pigment of interior feet in keeping with the eco-friendly situations of the habitat, for instance prey accessibility, aquatic temperature, and different resident circumstances.
This invertebrate (Scapharca subcrenata) is economically and ecologically very significant bivalves in the Korean peninsula. Even so, these types of invertebrates, those are known chief reproductively (Lee, 1998; Kim et al., 2008), ecologically (Kim et al., 2019), as well as bacteriologically (Shin et al., 2016) are not heritably and/or molecular-biologically studied like other crustaceans. There is a demand to apprehend the innate traits and structure of this invertebrate group with the aim of estimating surely the correctly heritable benefit.
PCR-fundamental examination approaches have been employed to pursuit the inherited characters of several living entities (Zhou et al., 2000; García et al., 2004; Liu et al., 2004; Yoon & Kim, 2004; Yan et al., 2005; Gelin & Souty-Grosset, 2006). The genomic markers particular to the lines, the strains, the species, the genera or the geographical groups have been utilized for the perception of entities, linkage, paternity testing, forensic medicine and mutation detection (Koh et al., 1999; Esselman et al., 2000; Kim & Choi, 2003; Dixon et al., 2004; Araneda et al., 2005; Yamazaki et al., 2005; Godhe et al., 2006; Kim et al., 2006; Yoon & Kim, 2010; Kang & Yoon, 2013; Yoon, 2019; Yoon, 2020). The author carried out clustering scrutiny to disclose the Euclidean genetic distances (GDs) of intra- and between-group of Scapharca subcrenata from Yeosu of the Korea.
MATERIALS AND METHODS
A specimen of 22 invertebrate (Scapharca subcrenata) individuals was obtained from a foreshore of Yeosu city of Korea. DNA analysis should be fulfilled as stated by the separation and extraction methods as before-mentioned by Yoon & Kim (2004). PCR abstraction was executed on DNA specimens of foot muscle tissues gathered from entities using five oligonucleotides polymers (ON-polymers). The lysis buffer Ⅰ was mixed to the specimens and this rinsing was repeated several times to clean the specimens. The density of the separated genomic DNA (gDNA) was analyzed grounded on the optical density at 260 nm by means of a spectrophotometer (Beckman Coulter, Buckinghamshire, UK). The pellets were re-suspended in ultra-pure distilled liquid and then used or frozen at −79°C until needed.
Expansions were fulfilled in a programmable thermal cycler (Perkin Elmer Cetus, Norwalk, CT, USA) by electrophoresis in 1.4% agarose gels, with PCR marker (Sigma-Aldrich Chemicals, Milwaukee, WI, USA) as DNA molecular weight marker and exposed by staining with ethidium bromide (EtBr) (Oh & Yoon, 2014). PCR upshots were refined using PCR Purification Kit (Qiagen Korea, Seoul, Korea) monitoring the producer’s procedures. The electrophoresed agarose gels were made lighter by ultraviolet (UV) treatment, and the gels were taken a picture operating a photographing apparatus (PECA, Beloit, WI, USA). Five ON-polymers, OPD-01 (5’-ACCGCGAAGG-3’), OPD-02 (5’-GGACCCAACC-3’), OPD-08 (5’-GTGTGCCCCA-3’), OPD-09 (5’-CTCTGGAGAC-3’), and OPD-15 (5’-CATCCGTGCT-3’) were shown to make the loci unique to each clam population (LUECP) and Loci shared by the two clam populations (LSTCP) which could be normally estimated.
Similarity matrix (SM) involving bandsharing (BS) grades between unlike entities in the two invertebrate groups was created in company with formula of Yoke-Kqueen & Radu (2006). Numerical scrutiny products such as Euclidean GDs, means and standard errors (SEs) were assessed by treating the hierarchical clustering platform Systat version 10 (SPSS, Chicago, IL, USA).
RESULTS AND DISCUSSION
At this point, the intricacy of the banding shapes fluctuated theatrically amid the ON-polymers from the shellfish di-group, as shown in Fig. 1. Especially, the banding forms were different largely from 550 bp to 850 bp between two clam groups. Particularly, DNA profiles with more fragments were observed in 300 bp than any other molecular sizes in two groups (Fig. 1). Five ON-polymers were used producing a total of 110 LUECP in group one and 132 in group two, respectively, varying in volume of DNA FRs from greater than almost 50 to not more than 1,050 bp, as demonstrated in Table 1. The higher FR amounts (>1,050 bp) are not noticed in the Scapharca subcrenata di-group. The DNA bands acquired by the five ON-polymers ranged from 200 to 1,500 bp in the discus (Symphysodon spp.) (Koh et al., 1999). Sixty-one bands varying from 200 bp to 1,900 bp were explicitly calculated in the ectoparasitic caligid (Lepeophtheirus salmonis) (Dixon et al., 2004). The amounts of the DNA FRs also fluctuated broadly, from 90 to 2,400 bp in Gracilaria vermiculophylla and G. chorda (Kim & Yoon, 2018). Yoon (2018) also reported that the greater FR sizes (>1,100 bp) are not detected in the razor shellfish (Solen corneus) bi-group.
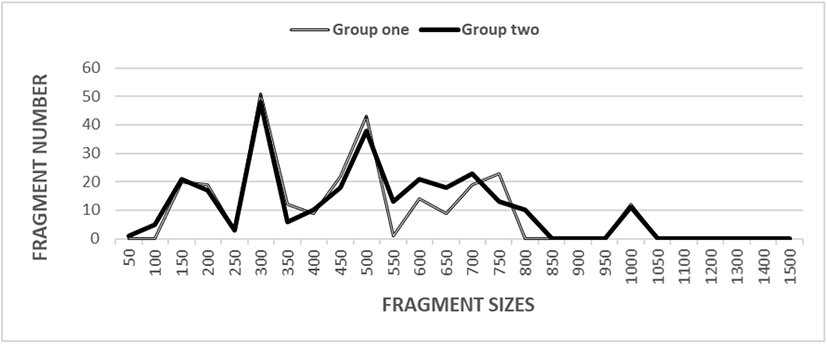
BS, bandsharing; SM, similarity matrix.
In this study, the BS grade between entity no. 04 of the group one and no. 09 of the group one was 0.478, which was the smallest observed, as displayed in Table 1. The BS grade between individuals’ no. 06 and no. 08 was 0.921 that was the utmost grade in the Scapharca subcrenata groups. In the present study, the five ON-polymers OPD-01, OPD-02, OPD-08, OPD-09 and OPD-15 made number of LUECP and number of LSTCP in two Scapharca subcrenata groups, as demonstrated in Table 2. The ON-polymer OPD-01 produced 33 loci LUECP, which were defining each population, almost 300 bp, 450 bp, and 500 bp, in the group one. Also, the 33 LUECP in the shellfish specimen made by the OPD-09 were approximately 300 bp, 500 bp, and 900 bp in size. Strangely, the ON-polymer OPD-15 recognized 22 LSTCP, dissimilar FRs of sizes 300 bp, which were corresponding in all specimens. The median number of LUECP was assorted and 1.2-fold greater in the shellfish group two than in the clam group one. As regards mean BS effects, entities in the clam group one (0.779±0.011) had a little higher BS grades than did entities from the shellfish group two (0.756±0.009) (p<0.05), as displayed in Table 3.
LUECP, loci unique to each clam population; LSTCP, loci shared by the two clam populations; ON-polymer, oligonucleotides polymer.
Each rate is an outcome of tri-dissimilar trials.
BS, bandsharing; SE, standard error; SM, similarity matrix.
| Group | Group one | Group two |
|---|---|---|
| Group one | 0.779±0.011a | 0.687±0.006b |
| Group two | - | 0.756±0.009ab |
In the present study, the entities of the shellfish group one are not tightly assembled with other entities of the invertebrate group two (Fig. 2). The PDG acquired by the five ON-polymers indicates two inherent groups: constellation I (SUBCRENATA 01, 09, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, and 22) and constellation II (SUBCRENATA 02, 03, 04, 05, 06, 07, 08, and 10). Amid the twenty-two shellfish, the smallest GD that exhibited momentous molecular alterations was between entities shellfish no. 22 and no. 19 from the constellation II (GD=0.057), while the largest GD among the 22 entities that exposed substantial molecular alterations was between entities clam no. 02 and clam no. 01 (GD=0.422). So, the GD (0.422) of this shellfish (SUBCRENATA 02 and 01) is 7.41-fold innately distinct to the GD (0.057) of the other shellfish (SUBCRENATA 22 and 19).
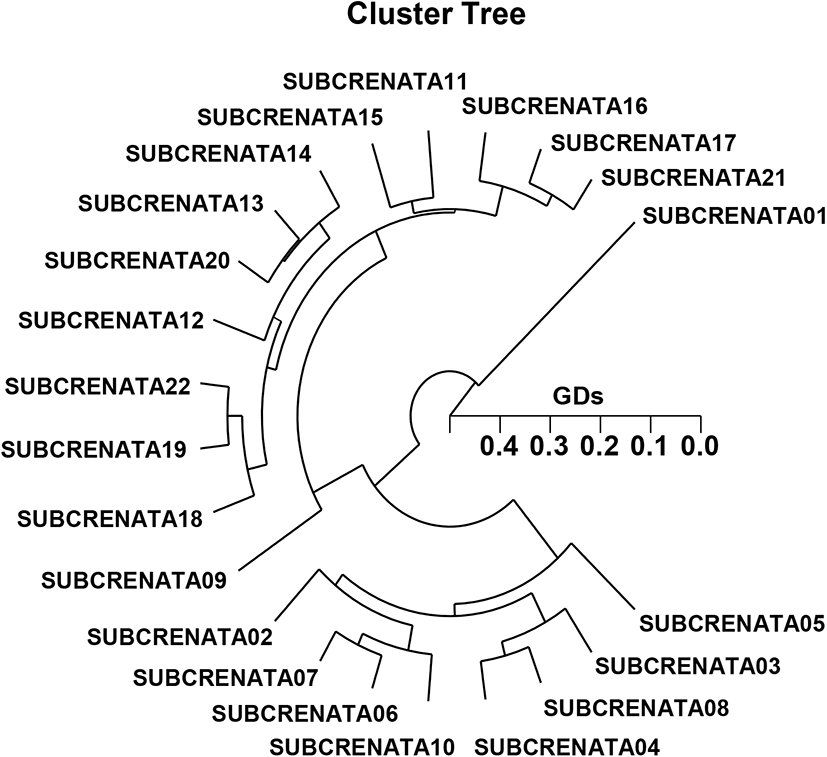
Above-mentioned, a PDG revealed close or distant relationships between entity structures amid bare figures and farmed varieties of discus (Symphysodon spp.) (Koh et al., 1999). Yoon (2018) reported that cluster analysis, which indicated two inherent constellations, revealed a relationship between the entities within two shellfish groups comparatively speaking other invertebrates. In the other shellfishes, cluster enquiry of the pairwise group matrix, created from heritable documents, indicated that geographically close groups be disposed to accumulate collectively in the blacklip abalone (Huang et al., 2000). S. subcrenata bi-group can be obviously categorized by gDNA-grounded processes.
The aptitude of ON-polymers augmented DNAs to determine unique markers for line, strain, species, genus, and group proof of identity in life entities (Koh et al., 1999; Esselman et al., 2000; Huang et al., 2000; Kim et al., 2000; Dixon et al., 2004; García et al., 2004; Araneda et al., 2005; Yamazaki et al., 2005; Yan et al., 2005; Gelin & Souty-Grosset, 2006; Godhe et al., 2006; Kim et al., 2006; Yoon & Kim, 2010; Kang & Yoon, 2013; Oh & Yoon, 2014; Kim & Yoon, 2018) has also been well approved. Great point of an important GD between two Scapharca subcrenata groups showed this research process is one of the most appropriate apparatuses for biotechnological DNA studies on entities and units of other life beings (Koh et al., 1999; Zhou et al., 2000; Kim & Choi, 2003; Dixon et al., 2004; Liu et al., 2004; Araneda et al., 2005; Godhe et al., 2006; Yoon, 2019; Yoon & Kim, 2010; Kang & Yoon, 2013; Yoon, 2020).
