INTRODUCTION
Krüppel-like factors (KLFs) encode C2H2-type zinc finger proteins that regulate various cellular functions, such as proliferation, differentiation, and apoptosis in the development (Black et al., 2001; Subramaniam et al., 2010). In mammals, 14 KLF genes have been identified as transcription factors; these belong to the Sp1 superfamily (Black et al., 2001). Human KLF10 was initially isolated from fetal osteoblasts treated with tumor growth factor-β (TGF-β); thus, its original name was TGF-β-inducible gene 1 (TIEG1) (Subramaniam et al., 1995). Human KLF11 (originally termed TGF-β-inducible gene 2) was identified by BLAST using the EST database for rat TIEG sequences (Cook et al., 1998). Overexpression of either KLF10 or KLF11 caused inhibition of cell proliferation and induction of apoptosis (Subramaniam et al., 2010). Mouse KLF10 knockout (KO) caused an increase in the predisposition of epidermal tumorigenesis and development of epidermal inclusion cysts after 7,12-dimethylbenz(a)anthracene and 12-O-tetradecanoylphorbol-13-acetate (DMBA/TPA) treatment (Song et al., 2012). Although the increased number of BrdU-positive cells in KLF10 KO mice supports the hypothesis that cell proliferation is inhibited by human KLF10, the relation between tumorigenesis in KLF10 KO mice and apoptosis in human KLF10-overexpressing cells remains unclear. As KLF10 KO mice exhibited a lower incidence of tumors during DEN-induced hepatic carcinogenesis (Heo et al., 2015), we suspect that a loss-of-function analysis in another model animal may aid understanding of the function of KLF10.
The Zebrafish Klf family is subdivided into four groups, consistent with the classification used for their mammalian homologs (Xue et al., 2015). Zebrafish Klf9, Klf10, Klf11a, Klf11b, Klf13, and Klf16 belong to members of group 4, which may facilitate transcriptional regulation by interacting with putative binding sites of Sin3A (Xue et al., 2015). Although a previous study showed that the respective losses of zebrafish Klf3 (group 3) and zebrafish Klf6a (group 2) led to an increased number of erythroid progenitor cells (EPC) and a smaller number of erythrocytes (Xue et al., 2015), these observations may be insufficient for a comprehensive analysis of the KLF family during embryogenesis in vertebrates. Moreover, they cannot provide new insights into the functions and mechanisms of KLF family during maintenance of cell homeostasis.
In the present study, we attempted a loss-of-function analysis of zebrafish homologs of human Klf10 by using antisense MO. Zebrafish Klf11a and Klf11b were isolated and characterized as homologs of human KLF10. Investigation of zebrafish Klf11b function in maintenance of cell homeostasis showed possible interactions between klf11b and p53. Our findings suggest that the relationship between Klf11b and p53 may play an important role during embryonic development in zebrafish.
MATERIAL AND METHODS
Zebrafish were maintained as described by Yeo et al. (2004). The embryos were staged in accordance with the method of Kimmel et al. (1995). AB* wild-type and Tg[islet1:GFP] (Higashijima et al., 2000) were used.
The sequences of zebrafish klf11a (NM001044941), klf11b (NM001077604), EF1a (NM131263), p53 (U60804), Bax (BC055592), and caspase 3 (NM131877) were identified in GenBank. Total RNA of Con-MO-, klf11a-MO-, and/or klf11b-MO-injected embryos were employed for RT-PCR. PCR was performed using the primers as described in Table 1. PCR products, so called cDNA, were subcloned into the pGEM-T vector, and then verified by sequencing.
Whole-mount in situ hybridization was performed as described by Yeo et al. (2004). Antisense riboprobes were transcribed from cDNAs for zebrafish klf11a, klf11b, p53, Bax, and caspase-3. Photos were taken with a differential interference contrast microscope (Axiopan2; Carl Zeiss, Oberkochen, Germany).
Immunohistochemistry was performed as described by Yeo et al. (2004). Anti-phospho-histone H3 was used as the primary antibody (Upstate). AlexaFluor 568-conjugated goat anti-rabbit IgG was used as secondary antibody (Molecular Probes). Photos were taken with a confocal laser scanning microscope (LSM700; Carl Zeiss).
Cell transplantation was performed as described by Jung et al. (2012). Cells from klf11b-MO- and fluorescein-dextran-injected embryos, as well as from klf11a-MO- and rhodamine-dextran-injected embryos, were used as donors. Cell transplants were performed at the mid-blastula stage. At 10 h after transplantation, embryos were fixed with 4% PFA and mounted using Vectashield with DAPI (Vector Labs). Confocal images were taken at the 15-somite stage with a confocal laser scanning microscope (LSM700; Carl Zeiss).
Microinjection was performed as described by Yeo et al. (2004). Spliced klf11a-MO (5’-TAG AGT CAC TCA CCA GTG ATG AGA G-3’; Gene Tools, Philmath, OR, USA), 4-bp mismatched klf11a control-MO (5’-TAG AGA CAC TGT CCA GTC ATG AGA G-3’; Gene Tools), spliced klf11b-MO (5’-GGT CAT GCA CTG CAG GTA AAC AGG A-3’; Gene Tools), 3-bp mismatched klf11b control-MO (5’-GGT CAA GCA CAC CAG GTA AAC AGG A-3’; Gene Tools), and p53-MO (5’-GCG CCA TTG CTT TGC AAG AAT TG-3’; Gene Tools) (2–4 ng) were injected into fertilized zebrafish eggs.
Total RNA was extracted from embryos at the 1-cell stage, as well as at 6-, 12-, and 24-hpf using TRIzol™ reagent (Invitrogen, Carlsbad, CA, USA). Their isolation was performed as described by manufacturer. Total RNA of Con-MOs-, klf11a-MO-, and/or klf11b-MO-injected embryos were used for Northern blot analyses. DIG-labelled zebrafish klf11a, klf11b, EF1a, p53, Bax, and caspase-3 were used as probes. Anti-Dig secondary antibodies were detected with an ECL kit (KPL, Boca Raton, FL, USA).
The TUNEL assay was performed as described in Song et al. (2012). Zebrafish embryos injected with Con-MOs or klf11b-MO were fixed with 4% PFA overnight and then dehydrated with methanol. Apoptotic cells were detected using the In Situ Cell Death Detection Kit, in accordance with the manufacturer’s instructions (Roche, Basel, Switzerland).
RESULTS
Two zebrafish homologs of human KLF10 were identified and characterized in silico using GenBank and the Sanger zebrafish genome assembly (www.ensembl.org). The predicted amino acid (AA) sequence of human KLF10 had approximately 54% and 49% similarity to predicted sequences of zebrafish Klf11b and Klf11a, respectively, whereas the predicted sequence of human KLF11 had approximately 57% and 57% similarity to predicted sequences of zebrafish Klf11a and Klf11b, respectively. Therefore, zebrafish Klf11a was classified as a homolog of human KLF11, while zebrafish Klf11b was classified as a homolog of human KLF10.
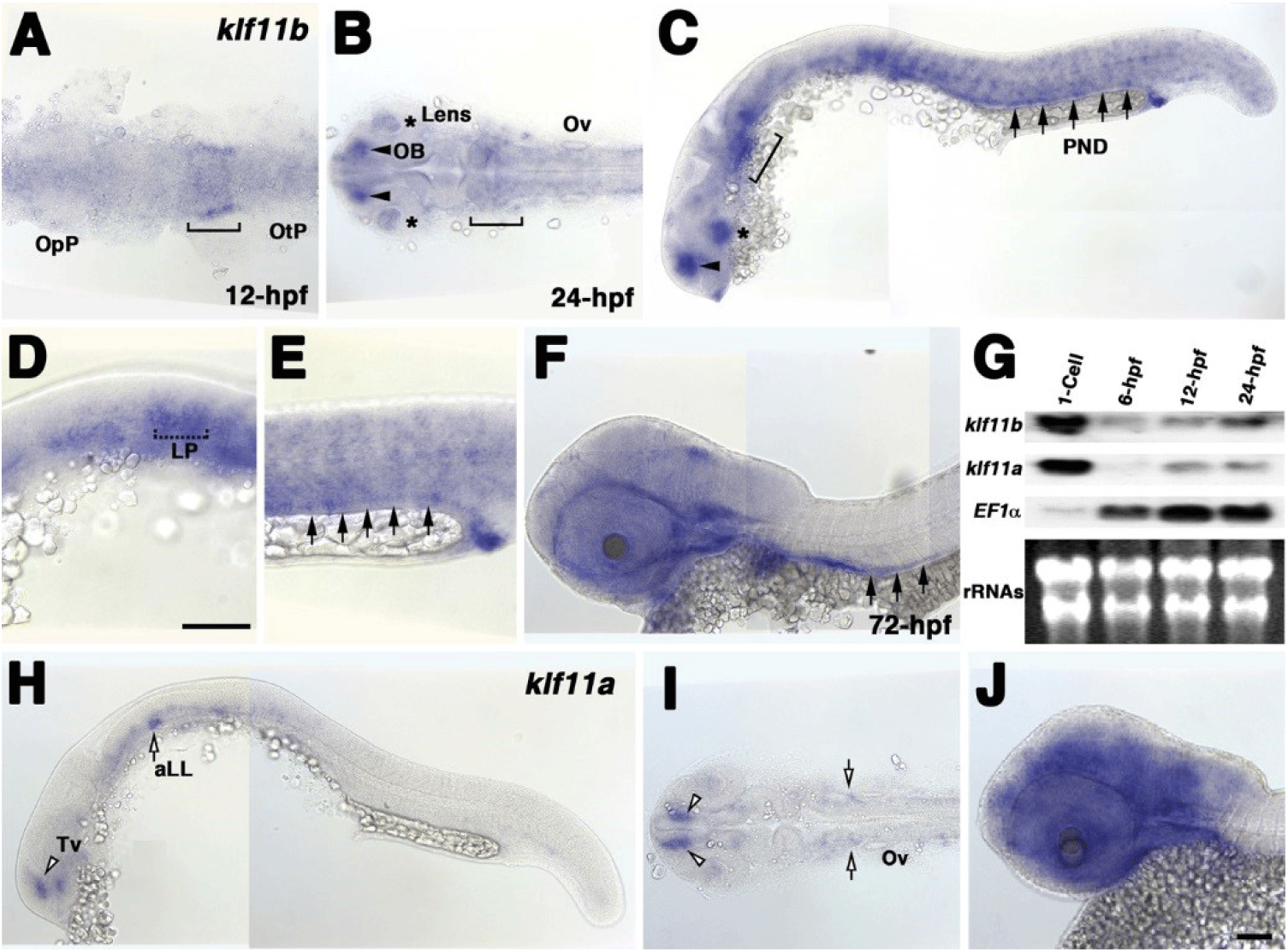
To examine the spatial and temporal expression of zebrafish homologs of human KLF10/11, we performed whole-mount in situ hybridization using klf11a and klf11b genes at various stages (Fig. 1). A very low level of klf11b mRNA was detected ubiquitously in zebrafish embryos at the gastrula stage (data not shown). At 12 h post-fertilization (hpf), mRNA expression of klf11b was concentrated in the hindbrain primordia of the neural plate (Fig. 1A). This expression in the neural plate was maintained in the hindbrain at 24 hpf (Fig. 1B). At this stage, klf11b mRNA was highly expressed in the olfactory bulb, lens, and somites, while weaker expression levels were detected in the nervous system (Figs. 1B and C). Transcripts of klf11b were also present in the pronephric duct (PND) and lateral primordia (Figs. 1C–E). At 72 hpf, transcripts of klf11b were consistently observed in the PND, with weaker expression levels seen in the nervous system (Fig. 1F). Northern blot analysis of zebrafish klf11 homologs showed maternal expression of their transcripts (Fig. 1G). Although transcripts of klf11a were difficult to detect by whole-mount in situ hybridization until 12 hpf, Northern blot analysis showed that klf11a mRNA was present at low levels from 12 hpf through 24 hpf (Fig. 1G). In contrast to the relatively broad expression of klf11b at 24 hpf, klf11a was expressed in spatially restricted domains of telencephalon, as well as in the anterior lateral line at low levels (Figs. 1H and 1I). At 72 hpf, transcripts of klf11a were persistently observed in telencephalon, with strong expression in the nervous system (Fig. 1J).
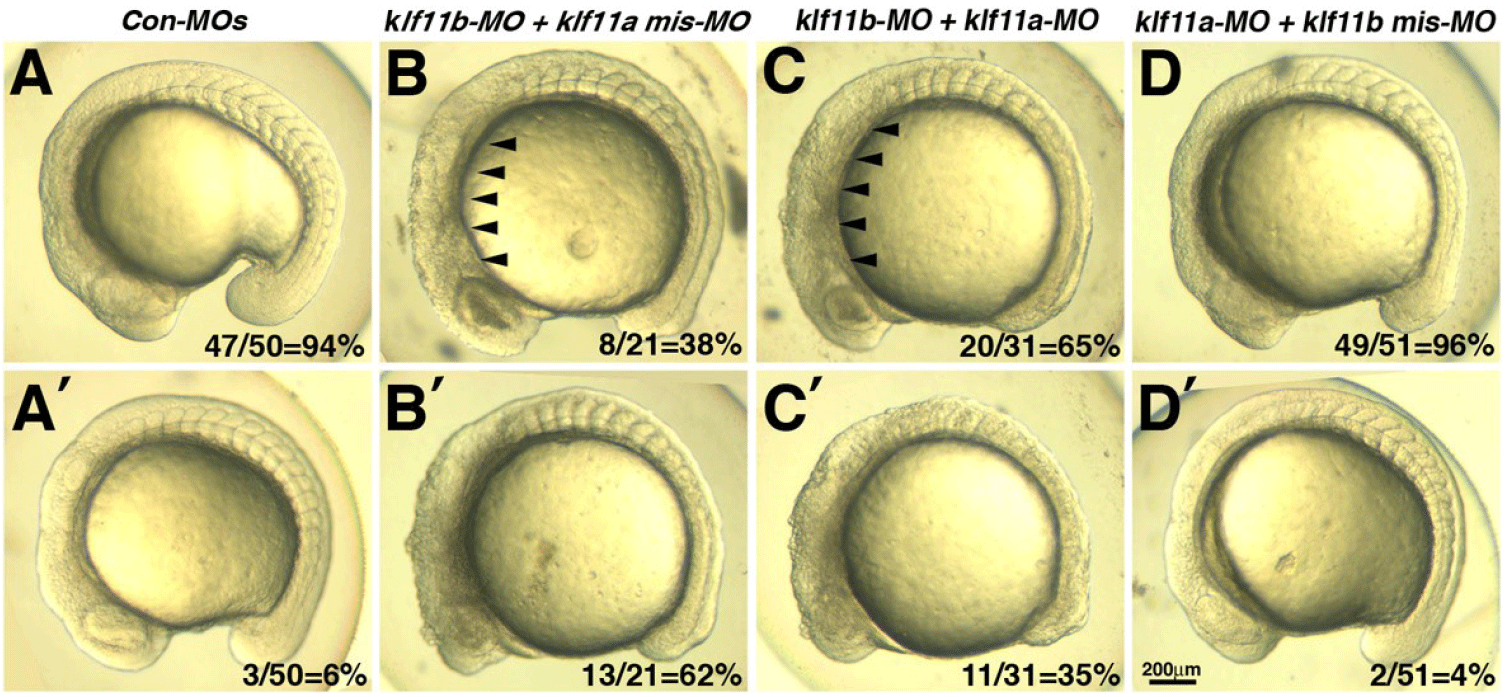
Our observation that transcripts of both klf11a and klf11b are maternally expressed suggested that they may be involved in control of early embryogenesis. As an initial step in analysis of the role of zebrafish homologs of human KLF10/11, we attempted to knock down klf11a and klf11b by injecting their MOs (ATG-MOs, which bind to a sequence on, or close to, the ATG start site of mRNA) into the embryos. A mixture of ATG-MOs caused severe developmental defects that interfered with analysis of morphological abnormalities after the gastrula stage (data not shown). Thus, we designed splice-site MOs to inhibit zygotic transcripts (Fig. 2). Reverse-transcription (RT)-PCR showed that embryos injected with 4 pg of klf11b-MO or klf11a-MO had abnormal amplicons of 1,728 base pairs (bp) or 1,721 bp at the 15-somite stage, respectively (Fig. 2A). Three bp-klf11b mismatch-MO was used as a control, and embryos injected with this MO did not generate alternate transcripts (Lane 3 in Fig. 2A). However, 4 bp-klf11a mismatch-MO slightly blocked pre-mRNA splicing of klf11a (Lane 2 in Fig. 2A). Abnormalities could be observed in embryos injected with a 4-pg mixture of klf11b-MO and klf11a mismatch-MO at the 15-somite stage, although these embryos developed normally through the end of gastrulation (Fig. 2C). By 16 hpf, all klf11 morphants could be separated into one of two groups: (i) moderate, characterized by cell death in the retina (Figs. 2C–2E); or (ii) severe, characterized by cell death in retina, as well as somites (Figs. 3B and 3C). The morphant phenotype was 54% moderate and 46% severe (n=52, Fig. 3). Embryos injected with 4 pg of klf11b mismatch-MO and klf11a mismatch-MO (Con-MOs) (Fig. 2B, n=50), as well as those injected with klf11a-MO and klf11b mismatch-MO (Fig. 2D, n=51), developed normally until 16 hpf (Fig. 2, Fig. 3).
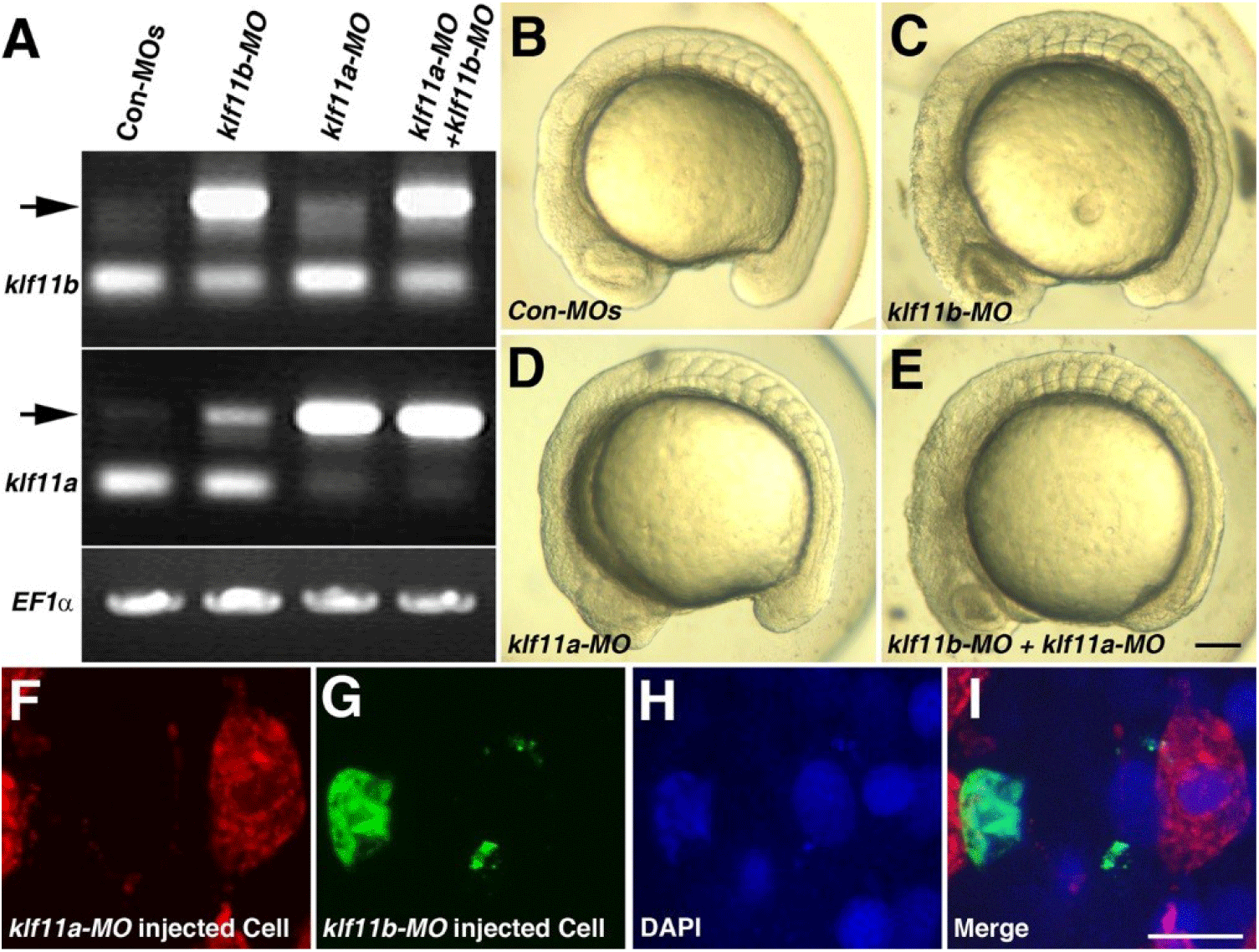
To assess whether Klf11a and Klf11b function in a cell-autonomous manner, we performed cell transplantation experiments (Figs. 2F–2I). At 3.5 hpf, cells from donor embryos that had been injected with klf11b-MO and FITC-dextran were mixed with cells from embryos that had been injected with klf11a-MO and rhodamine-dextran, and then transferred into wild-type embryos (Fig. 2I). At 16 hpf, klf11a-MO-injected cells were normally developed (Fig. 2F), whereas atypical nuclei were observed in the green channel (Figs. 2G–2H). This observation suggested that cells injected with klf11b-MO might undergo cell death, and that Klf11b functions in a cell-autonomous manner.
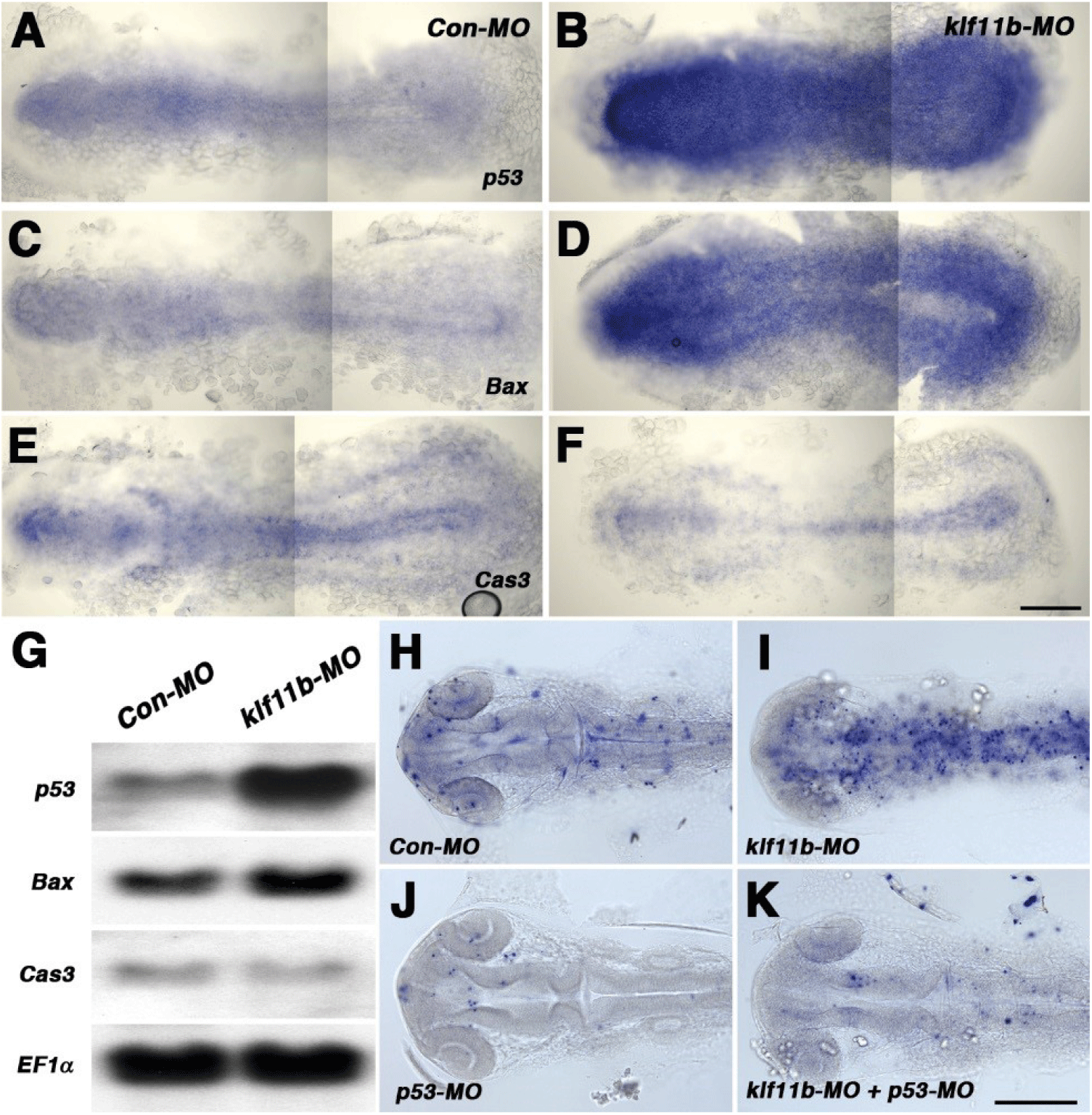
To assess embryos injected with klf11b-MO, we performed whole-mount in situ hybridization with p53, Bax, and caspase-3, all of which are involved in cell death in zebrafish (Fig. 4). Based on phenotypic analysis (Fig. 2C), embryos injected with klf11b-MO were fixed at the 5-somite stage (Figs. 4A–4F). Zebrafish p53 was ubiquitously expressed in Con-MO-injected embryos (Fig. 4A). However, its transcripts were dramatically increased in klf11b-MO-injected embryos (Fig. 4B). Compared to control embryos (Fig. 4C), the expression of bax, a direct target of p53, also increased (Fig. 4D). These data were consistent with the hypothesis that overexpressed Bax accelerates apoptotic cell death and counters the death repressor activity of Bcl-2 (Oltval et al., 1993). The expression of caspase-3 was examined at the 5-somite stage (Fig. 4E), which showed that overexpression of zebrafish caspase-3 induced apoptosis and served to modulate the pro-apoptotic signal during zebrafish development (Yabu et al., 2001). The expression level of caspase-3 remained unchanged, or slightly reduced, in the klf11b-MO-injected embryos compared to that in Con-MO-injected embryos (Figs. 4E and 4F). Northern blot analysis confirmed increased expression levels of p53 and bax, as well as unchanged expression levels of caspase-3, in the klf11b-MO-injected embryos (Fig. 4G). These findings suggested that the phenotype of Klf11b knockdown embryos might be rescued by reduced expression of p53. To test this, we performed the TUNEL assay in klf11b-MO- and/or p53-MO-injected embryos at 26 hpf (Figs. 4H–4K). Apoptotic cells were observed in embryos injected with Con-MO (Fig. 4H, n=50), as well as in embryos injected with klf11b-MO (Fig. 4I, n=23). However, the number of apoptotic cells was dramatically reduced in embryos co-injected with klf11b-MO and p53-MO (Fig. 4K, n=34), as well as in embryos injected with p53-MO (Fig. 4J, n=29). MO-mediated knockdown of p53 in klf11b-MO-injected embryos rescued the apoptotic phenotype, implying that the function of Klf11b was equivalent to that of p53 or was upstream of p53 in the p53-dependent apoptotic pathway.
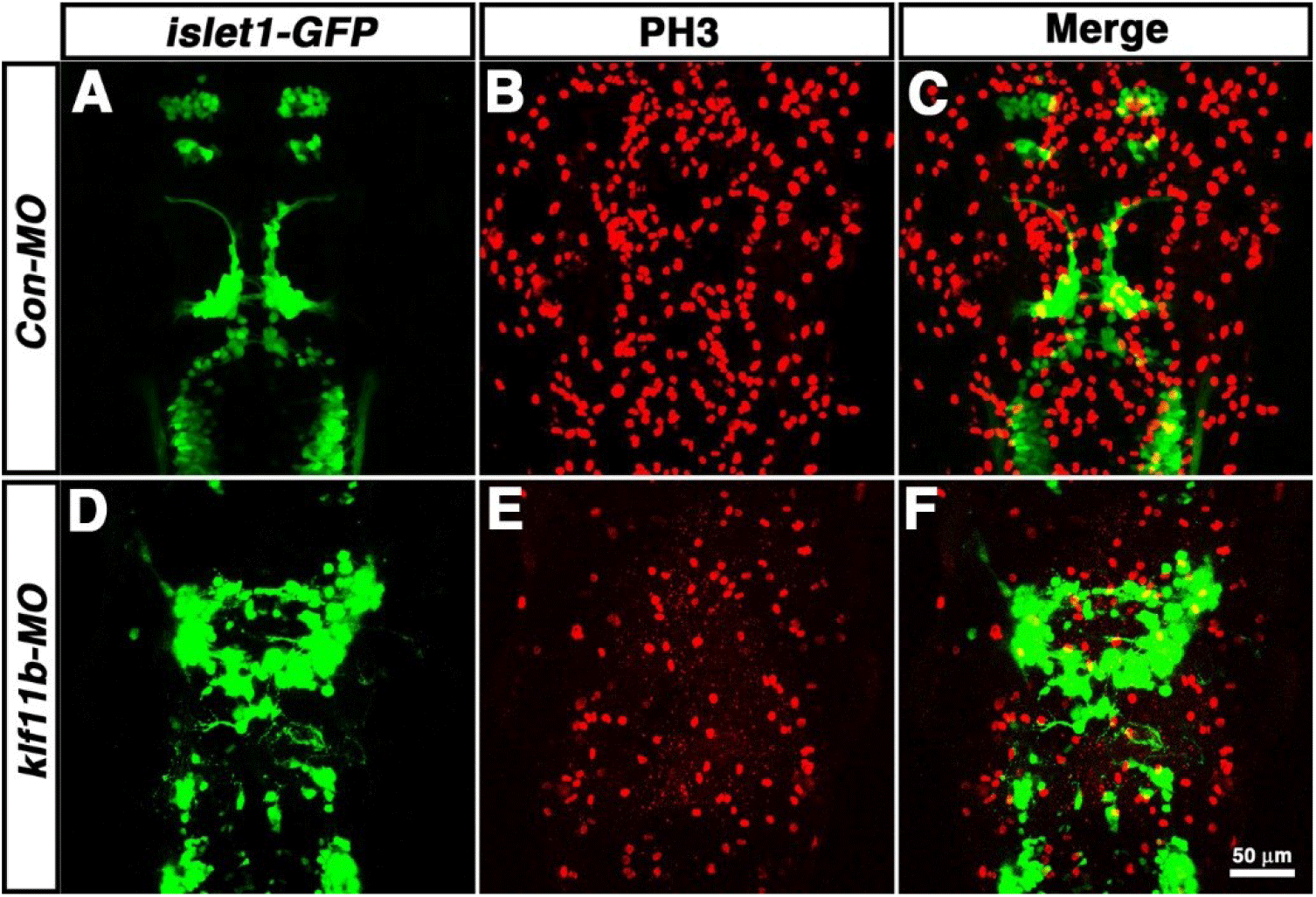
To determine which cell types are most strongly affected after knockdown of klf11b function, Tg[islet1-GFP] embryos were injected with klf11b-MO, and then labeled with an antibody that recognized phosphorylated histone H3 on Ser-10 (PH3), an established marker for chromosome condensation during mitosis (Fig. 5, Hendzel et al., 1997; Higashijima et al., 2000). In the Tg[islet1-GFP] line, GFP mRNA is expressed in a majority of cranial motor neurons, in a manner similar to that of islet-1 mRNA, beginning at 21 hpf (Higashijima et al., 2000). At 36 hpf, GFP-positive cranial motor neurons were well organized spatiotemporally in the central nervous system (CNS) of Con-MO-injected embryos (Fig. 5A), whereas morphological and spatial disorganization of GFP-positive cells was observed in the CNS of klf11b-MO-injected embryos (Fig. 5D). Although the number of GFP-positive cells was difficult to determine, the number of PH3-positive cells significantly decreased in klf11b-MO-injected embryos (Fig. 5E; 122.3±9.3, n=6), compared to that in the Con-MO-injected embryos (Fig. 5B; 342.8±8.7, n=4). These observations implied that reduction of proliferating precursors may be caused by apoptosis.
DISCUSSION
We identified and characterized zebrafish Klf11b as a homolog of human KLF10. Structural comparisons showed that zebrafish Klf11 homologs are highly conserved, with respect to human KLF10 and KLF11. Both zebrafish klf11a and klf11b mRNA are maternally expressed. During zebrafish development, klf11a is expressed in the ventral telencephalon and anterior lateral line, while klf11b is highly expressed in the olfactory blub, lens, PNDs, lateral primordia, and somites. However, only klf11b transcripts can be detected at the gastrulation stage. This might be the cause of epiboly retardation in klf11b morphants, as well as the source of their degenerative phenotype after this stage. Although structural preservation of zebrafish Klf11 homologs strongly suggests conservation of KLF homolog functions during evolution, the function and expression according to specific spatiotemporal patterns may have varied among individual subtypes.
Structural alignment showed that three zinc-finger domains of human KLF10 had approximately 79% and 84% similarity to zebrafish Klf11a and Klf11b, respectively, whereas human KLF11 had approximately 88% and 93% similarity to zebrafish Klf11a and Klf11b, respectively. Although zebrafish Klf11b is classified as a homolog of human KLF10 based on comparison of the overall AA sequence, zinc-finger domains of zebrafish Klf11b showed greater similarity with those of human KLF11. This may be why zebrafish Klf11a and Klf11b were characterized as human KLF11 homologs. In the zebrafish genome assembly, another Klf homolog is present: Klf10, in chromosome 16. Human KLF10 and zebrafish Klf11b consist of 469 and 458 AAs, respectively, whereas zebrafish Klf10 only contains 218 AAs. Comparison of predicted AA sequences showed that the zinc-finger domain (approximately 132 AAs) of zebrafish Klf10 had approximately 69%, 71%, 68%, and 68% similarity to zebrafish Klf11a, zebrafish Klf11b, human KLF10, and human KLF11, respectively. However, neither whole-mount in situ hybridization nor RT-PCR could detect zebrafish Klf10 transcripts (data not shown), suggesting that zebrafish Klf10 might be a pseudogene.
Northern blot analysis has shown that human KLF10 (TIEG1) transcripts are highly expressed in skeletal muscle, and moderately expressed in heart, placenta, and pancreas, whereas KLF11 (TIEG2) mRNA is ubiquitously expressed, with the highest levels occurring in the pancreas and skeletal muscle (Subramaniam et al., 1995; Cook et al., 1998). Human KLF11 is more prominently expressed in the pancreas, whereas KLF10 is expressed at higher relative levels in skeletal muscle (Cook et al., 1998). In zebrafish, whole-mount in situ hybridization showed that only klf11b transcripts are present in muscle, although both klf11a and klf11b are highly expressed in the brain of 3-day-old embryos. Taken together with the AA sequence comparison, the most likely classification of zebrafish Klf11b is as a homolog of human KLF10.
In the present study, we showed that a reduction in the number of proliferative precursor cells may be the result of cell death in zebrafish Klf11b morphants, suggesting that such cell death may be mediated by p53. Biochemical analysis might elucidate that a KLF gene might be a negative regulator of p53 transcription. Based on these observations, we propose three models: first, zebrafish Klf11b may function as a transcriptional repressor, with or without a corepressor, by binding to Sp1 elements of the p53 gene; second, zebrafish Klf11b may function as a corepressor by binding to p53 protein; and finally, DNA-binding Klf11b may mask the function of p53 as a transcriptional activator. Although interaction of the corepressor mSin3A with an alpha-helical motif of Klf11/ Klf10 supports one of our hypotheses (Zhang et al., 2001), previous studies have shown that overexpression of human KLF10 in cancer cell lines induced apoptosis, in a manner similar to that of the mitochondrial pathway (Tachibana et al., 1997; Ribeiro et al., 1999). Additionally, transgenic overexpression of KLF11 has been shown to result in a decreased number of proliferative cells, increased rate of apoptosis, and reduction in the size of the pancreas (Fernandez-Zapico et al., 2003). In human leukemia cells, overexpression of human KLF10 promoted chemically induced apoptosis through the mitochondrial pathway, as represented by upregulation of Bax, downregulation of Bcl-2, release of cytochrome C, and activation of caspase-3 (Jin et al., 2007). These studies indicate that loss of KLF10 function could promote carcinogenesis (Tachibana et al., 1997; Ribeiro et al., 1999; Fernandez-Zapico et al., 2003). Previous study provides some support for this hypothesis, based on treatment with carcinogens such as DMBA and TPA (Song et al., 2012). However, KLF10-deficient mice demonstrated increased numbers of apoptotic cells in DMBA/ TPA-treated epidermis, and lower incidence of tumors, than wild-type mice, following DEN-induced liver tumorigenesis (Song et al., 2012; Heo et al., 2015).
Although human KLF6 is essential for hepatic carcinoma cells to evade p53-mediated apoptosis in vitro (Sirach et al., 2007), our study provides the first observation concerning the relationship between KLFs and p53 in vivo. To better understand the function of KLFs, further studies are needed to address their roles in apoptosis, as well as in tumorigenesis. First, KLF10 and KLF11 double KO mice could help determine the functions of these proteins. In humans, KLF10 and KLF11 exhibit approximately 84% similarity, and their expression pattern is very similar in adult human tissues (Cook et al., 1998). Based on the principal of genetic redundancy, KLF10 and KLF11 double KO mice may show functions of KLFs during gastrulation, as the expression of mouse KLF11 is unclear during early embryonic development. As described above, zebrafish klf11b is expressed at the gastrulation stage, whereas klf11a is not, and the apoptotic phenotype is observed after gastrulation. Second, co-immunoprecipitation with the KLF11b and p53 antibody as well as isolation and characterization of genes downstream of the KLF11b/p53 complex could aid in characterizing their functions. Finally, examination of p53 stability in the absence of mammalian KLF10 could help determine its function. Mungamuri and colleagues reported that USP7 deubiquitinase protected SUV39H1 from MDM2-mediated ubiquitination in the absence of a p53 stimulus (Mungamuri et al., 2016).
In summary, we have shown that Klf11b morphants undergo apoptosis, which may be mediated by p53. Zebrafish Klf11b morphants could be used in genetic screening for identification of unknown genes, as well as for identification of small molecules involved in chemically induced cell death. This model will greatly facilitate understanding of the homeostasis underlying cell viability, which is mediated by the interaction between KLF and p53.
