INTRODUCTION
Korean pen shell (Atrina pectinata) is one of economically outstanding large shell species in the West sea, fitting to the family Pinnidae. In the ecological bio-system, A. pectinata shell is mainly occupied in the seawater areas of the Korean peninsula, besides in some Asian Pacific districts in the China, Japan, Taiwan, and Indonesia. Pen shell shall be mainly found in the mud of the intertidal zone from 5 to 10 m, depth of water. Spawning period is from July to August. The body color of the shellfish is grayish-brunet or dark-yellowish. Distinctly, the dorsal surface of shell is routinely velvety and three-sided. The sizes of the pen shell is 250–300 mm, height 140–150 mm, width roughly 100 mm. The triangular-shaped pen shell is much larger than any other venerid clams in the Korean peninsula. Since shells are hours of darkness living beings, they settle under the gravels, filthy mud and pillars of the shallow marine (Min, 2001). Many explores have displayed that the change of aquatic temperature by earth warmth, population concentration, tide wall, thoughtless overfishing and environmental confusion are serious in the early larval development of this pen shell. Korean pen shell is one of marketable-scale and preferred crustaceans throughout the year.
Nevertheless their commercial and environmental appraisals, simply some records currently are remaining that are recognized as epidemiologically (Choi et al., 2017), reproductive ecologically (Yoo & Yoo, 1984), resource biologically (Kim et al., 2007), developmentally (Yoo et al., 1988; Lee et al., 2006; Chung et al., 2012), sitologically (Cha et al., 1995; Hwang et al., 2018), physiologically (Han et al., 2020), and molecular biologically (An et al., 2011) associated to other shellfish species. Accordingly, there is a criterion to appreciate the genetic traits and more statistics of this clam so as to appraise unerringly the accurately genetic suggestion. Most of all, the grouping probe of the genetic distance (GD) between populations/species/genera of quite a few finfish and shellfishes from the altered terrestrial locations has been achieved by dint of polymerase chain reaction (PCR) modus operandi is not many number (Diaz-Jaimes & Uribe-Alcocer, 2003; Wang & Li, 2004; Wasko et al., 2004; Nagarajan et al., 2006; Upadhyay et al., 2006; Fernandez-Tajes et al., 2007; Mahmoud et al., 2010; Kang & Yoon, 2013; Jo & Yoon, 2021).
This inspection tries to clarify the GDs and polymorphism indoors and amid pen shell collections. With the purpose of attainment, this inventor undertook clustering probes of pen shell (A. pectinata) in the West Sea of Korea.
MATERIALS AND METHODS
Muscle tissues were accumulated unconnectedly as of pen shell (A. pectinata) from Seosan of the Korean peninsula. PCR probe was carried out on DNA samples extracted from a total of 22 sample testers using seven oligonucleotides primers (OLIGO-primers). DNA extraction should be fulfilled on the word of the extraction procedures (Kang & Yoon, 2013). The concentration of the extracted genomic DNA (gDNA) was gauged by optical density at 260 nm by a spectrophotometer (Beckman Coulter, Buckinghamshire, UK). The DNA bits were incubation-dehydrated for 2 hrs, detained at −80°C until DNA probe and then liquefied in the TE buffer (10 mM Tris-HCl, pH 8.0; 1 mM EDTA).
Seven OLIGO-primers, for instance, OPD-01 (5’-ACCGCGAAGG3’), OPD-02 (5’-GGACCCAACC-3’), OPD-06 (5’-ACCTGAACGG-3’), OPD-08 (5’-GTGTGCCCCA-3’), OPD-09 (5’-CTCTGGAGAC-3’), OPD-15 (5’-CATCCGTGCT-3’) and OPD-18 (5’-GAGAGCCAAC-3’) were used to generate the unique loci shared to each pen shell team (ULSEPT) and quantity of loci shared by the binary pen shell teams (LSBPT) that could be clearly scored. Enlargement products were deployed by electrophoresis in 1.4% agarose gels (Bioneer, Daejeon, Korea) with Tris-borate buffer (0.09 M Tris, pH 8.5; 0.09 M borate; 2.5 mM EDTA). The molecular sizes were measured by assessment using 100 bp DNA ladder (Bioneer) as DNA molecular weight marker. The subsequent PCR probes were performed using Programmable DNA Thermal Cycler (MJ Research, Waltham, MA, USA). After electrophoresis, gels were stained and revealed by staining with EtBr, viewed with ultraviolet ray transilluminator, and then photographed by photoman direct copy system (PECA products, Beloit, WI, USA).
The ABS amounts were successively gauged by the presence/absence of expanded outcomes at the peculiar spots from mean fragment (FRA) reproducibility of the DNA outlines. Similarity matrix comprising BS extent between different individuals in the binary shellfish populations was created as stated by formula of Yoke-Kqueen & Radu (2006). A clustering tree was constructed using SM to create a hierarchical circular dendrogram (HCDG), which was expedited by the Systat version 10 (SPSS, Chicago, IL, USA).
RESULTS AND DISCUSSION
In current inquiry, gDNA disconnected from binary populations of Korean pen shell (A. pectinata) was amplified by PCR research. The trilateral SM for pairwise evaluation of BS grades of PSTs, ranged from 0.499 to 0.884 in the pen shell team A (PSTA) and 0.495–0.768 in the pen shell team B (PSTB), as shown in Table 1. The BS amount between entity’s no. 01 and no. 05 was the highest (0.884) between the binary PSTs. The BS grade between entities no. 02 and no. 12 was the lowest (0.377). Those major and minor FRAs were around from 50 bp to 2,000 bp in FRA size and comparable in all teams sampled from both teams, as shown in Fig. 1. In the present study, seven OLIGO-primers were used to create the DNA FRA size ranging from 50 to 2,000 bp. A whole of 61 loci were established extending in size from 400 to 2,000 bp in the razor clam, Ensis silique (Fernandez-Tajes et al., 2007). The figures of the DNA FRA size showed lower quantities than those presented by other researchers.
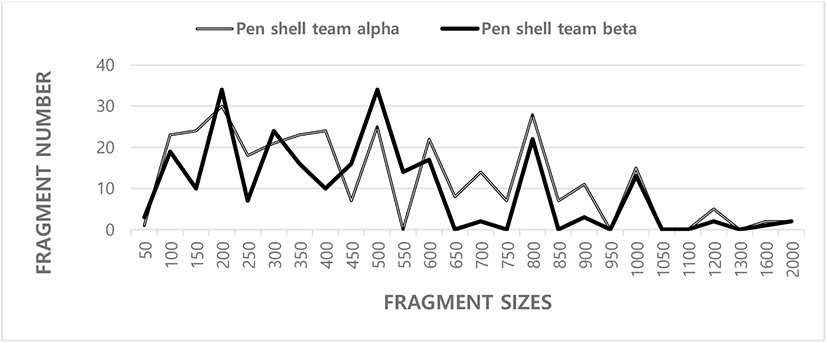
SM, similarity matrix; BS, band-sharing; PSTA, pen shell team A; PSTB, pen shell team B.
The seven OLIGO-primers were consumed to produce the quantity of ULSEPT and LSBPT (Table 2). 328 FRAs were identified in the PSPA, and 257 in the PSPB: 77 quantity of ULSEPT (23.48%) in the PSTA and 121 (47.08%) in the PSPB. 154 quantity of LSBPT, with an average of 22.0 per primer, were detected in the binary PSTs. 154 quantity of LSBPT, with an average of 22.0 per primer, were detected in the binary PSTs. The 22 ULSEPT in the PSTA tester were produced using the OPD-01 OLIGO-primer, approximately 500 bp and 1,000 bp in size. Surprisingly, the primer OPD-08 generated 22 ULSEPT (approximately 100 bp and 450 bp in size) that were shared with each team of PST that could be used to classify each team. Assessed independently, the average ULSEPT was higher in the PSPB than in the PSTA. On the other hand, the OPD-02 OLIGO-primer didn’t create 0 ULSEPT and 0 LSBPT, respectively. 212 polymorphic FRAs were gained from a total of 414 hydroid (Sertularia cupressina) colonies by means of 10 PCR primers used (Henry & Kenchington, 2004). The primer BION-80 detected 7 shared loci by the three venerid clam species, major and/or minor FRAs of sizes 500 bp, which were matching in all samples (Jeon & Yoon, 2015).
PSTA, pen shell team A; PSTB, pen shell team B; PSTs, pen shell teams; OLIGO-primers, oligonucleotides primers; ULSEPT, unique loci shared to each pen shell team; LSBPT, loci shared by the binary pen shell teams; FRAs, fragments.
The ABS grades of entities in the PSTA (0.685±0.011) were higher than in those originated from the PSTB (0.640±0.009) (p<0.05), as well-known in Table 3. Related discretely, the ABS grades of the entities PSTA were higher than those from PSTB. To discriminate, the ABS amounts of individuals in the Charybdis crab population A (CCPA) (0.575±0.014) were lesser than in those originated from the Charybdis crab population B (CCPB) (0.705±0.011) (p<0.05) (Yoon, 2022), and entities from Cyclina sinensis species (0.754) displayed higher BS amounts than did entities from Saxidomus purpuratus species (0.607) (p<0.05) in the three venerid shellfish species (Jeon & Yoon, 2015).
| Team | PSTA | PSTB |
|---|---|---|
| PSTA | 0.685±0.011a | 0.578±0.006c |
| PSTB | - | 0.640±0.009b |
Each amount is a result of three disparate experiments.
BS, band-sharing; SM, similarity matrix; PSTA, pen shell team A; PSTB, pen shell team B.
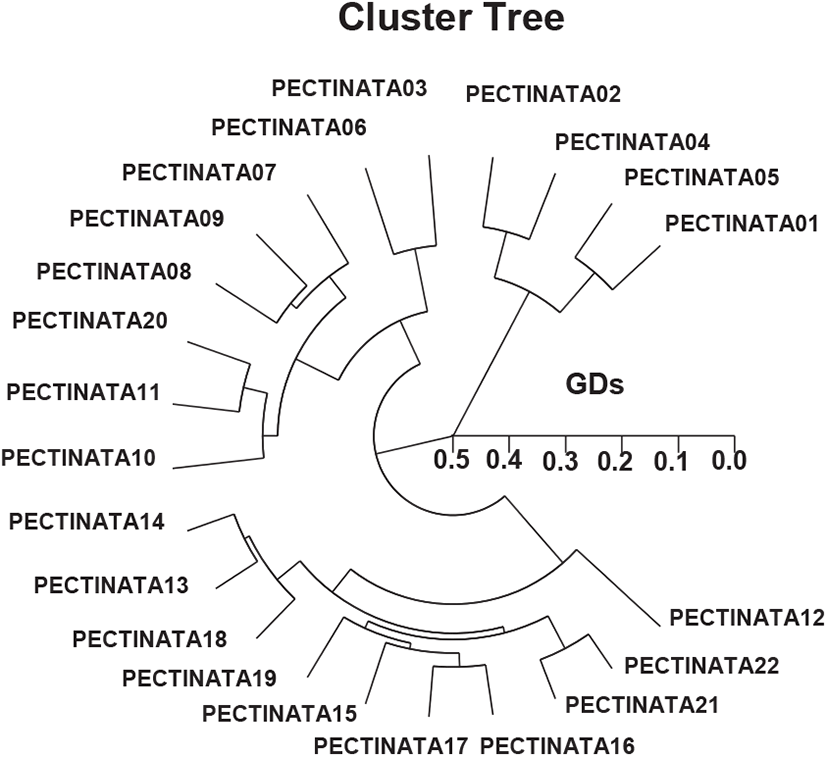
This author reported that complete linkage cluster method, which indicated binary genetic groupings, and the dendrogram revealed a close relationship between the identities within binary PSTs. The HCDG achieved by the seven OLIGO-primers denotes binary genetic groups: group alpha (PECTINATA 01, 02, 03, 04, 05, 06, 07, 08, 09, 10, 11, and 20), group beta (PECTINATA 12, 13, 14, 15, 16, 17, 18, 19, 21, and 22), as presented in Fig. 2. The highest GD indicating noteworthy molecular difference was between individuals PECTINATA no. 06 and PECTINATA no. 04 (0.498). Toro et al. (2004) insisted the amounts of Nei’s unbiased GD, D (0.030–0.107), among populations were small, despite the large geographic separation in the Chilean blue mussel Mytilus chilensis. The rates of the pairwise evaluations of equitable GD between the populations of the Indian major carp (Catla catla) from the pooled records for the four primers, ranged from 0.025 to 0.052 (Islam et al., 2005). They informed that the Padma and the Jamuna populations were divided from each other with the lowest GD (=0.025). The amounts of Nei’s GD were small among the Spanish and Portuguese populations (0.051–0.065), and high between these and the Irish populations (Fernandez-Tajes et al., 2007). Cluster and principal coordinate probes supported these endings. A mantel test performed between geographic and GD matrices showed a significant correlation (r=0.84, p<0.05), suggesting an isolation by distance process.
This modus operandi might help define the inquisitive pointers related to the lines, species, genera, and geographic populations identification in teleost (Garcia et al., 2004; Araneda et al., 2005; Siti Azizah et al., 2005; Upadhyay et al., 2006), and crustaceans (Esselman et al., 2000; Huang et al., 2000; McCormack et al., 2000; Dixon et al., 2004; Henry & Kenchington, 2004; Fernandez-Tajes et al., 2007; Mahmoud et al., 2010; Yoon, 2022). By way of this research, it is possible a little to contribute to expanding the farming of pen shells, maintenance of clam species, protection of the natural environment, and conservation of ecosystems.
