INTRODUCTION
The deleted in azoospermia like (DAZL) gene belongs to the deleted in azoospermia (DAZ) gene family. DAZ gene family comprises DAZ (DAZ1, DAZ2, DAZ3, and DAZ4), deleted in azoospermia like (DAZL), and BOLL (boule homolog RNA binding protein, BOULE). These genes encode RNA-binding proteins (Lin & Page, 2005; Yu et al., 2008; Zagore et al., 2018). As an autosomal gene, the DAZL gene is located on chromosome 3 (3p24.3) in humans and chromosome 17 (25.89 cM) in mice. It is widely recognized as a highly conserved and important regulator in germ cell development. DAZL gene is identified through homology with the Y-chromosome-linked DAZ gene. In humans, polymorphisms or mutations in DAZL have been associated with male infertility, such as azoospermia and oligozoospermia (Reijo et al., 1995; Reynolds & Cooke, 2005). In the mouse model, loss of the DAZL gene leads to complete germ cell depletion, with affected germ cells failing to enter meiosis and undergoing apoptosis (Lin et al., 2005). DAZL gene is now known to be expressed in both sexes and at various stages of germ cell differentiation (Cooke et al., 1996). It has been revealed that DAZL involves ovarian insufficiency and premature ovarian failure (Liu et al., 2024).
Unlike DAZ gene, which is limited to the Y chromosome in primates, DAZL is conserved across vertebrates, including humans, mice, frogs, and zebrafish (Reijo et al., 1995). DAZL gene encodes DAZL protein, an RNA-binding motif containing DZA RNA binding protein family. Interaction with RNA supports the role of DAZL protein as it enhances or suppresses translation depending on the developmental context (Yan et al., 2022; Yang et al., 2024).
Given the central role of DAZL in gametogenesis, understanding translational regulation and its mechanisms is critical for advancing reproductive biology and developing clinical interventions for infertility. In addition, recent studies also suggest other possible biological roles. In here, the DAZL gene structure and highlighting emerging areas of research that extend beyond the germline will be summarized.
DELETED IN AZOOSPERMIA LIKE GENE ANATOMY AND ITS EXPRESSION
DAZL gene is a protein coding gene (transcription start site region 148594, Tssr148594), single copy both in human and mouse. It maps to 3p25 of chromosome 3 in humans and chromosome 17 in mice (Seboun et al., 1997). The DAZL gene arose in the early vertebrate lineage from the BOLL gene, which is the ancestral gene conserved from flies to humans. DAZ arrived on Y chromosome during primate evolution from the ancestral autosomal DAZL gene (Xu et al., 2001; Yu et al., 2008).
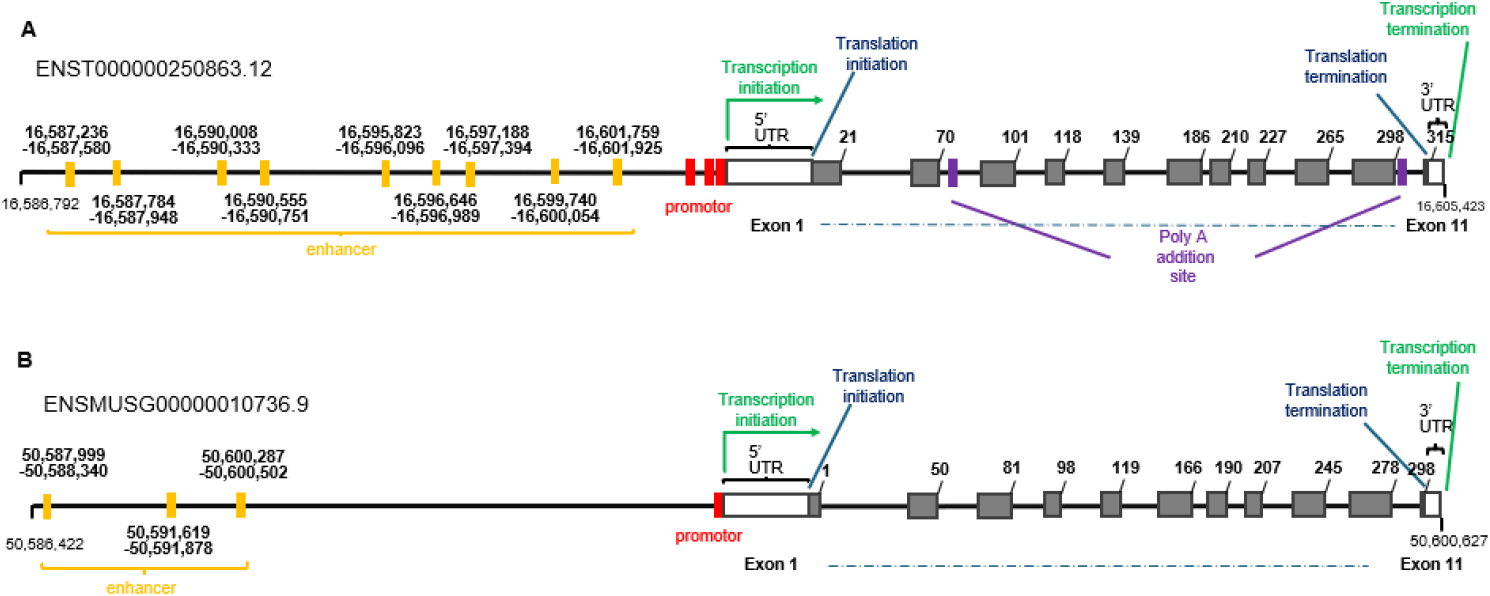
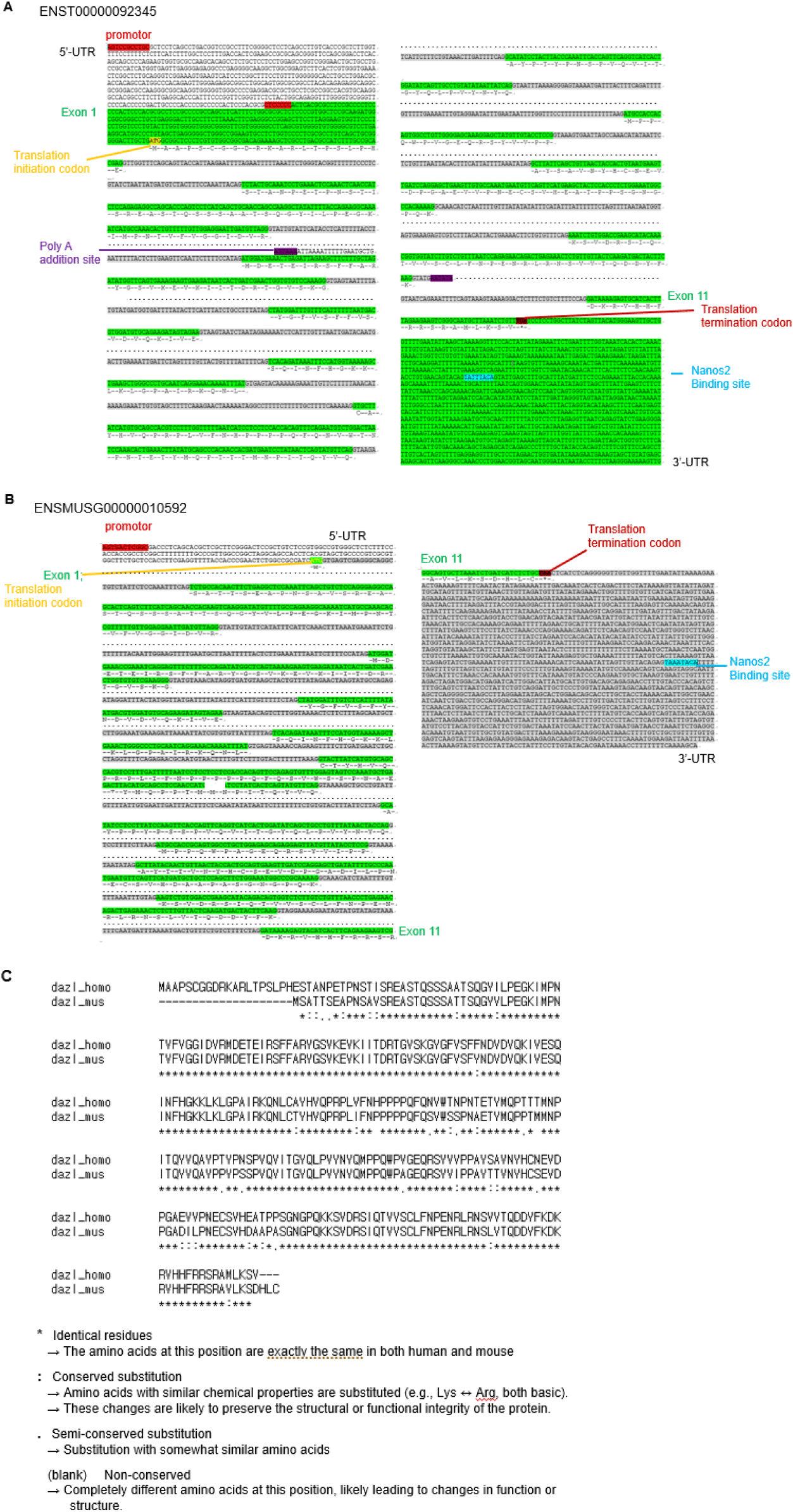
DAZL has many synonyms: Dazla, Daz-like, Tpx-2, Tpx2 in mouse, and DAZLA, DAZH, SPGYLA, MGC26406, DAZL1 in human (Baldarelli et al., 2024; NCBI, 2025). In human and mouse, DAZL is constructed with 11 exons and 10 introns, with the transcription start codon at exon 1 and the stop codon at exon 11. 5’-untranslated region (UTR) and 3’-UTR locate each translation initiation codon and translation terminal codon, respectively. Enhancers are located upstream of the promoter (Fig. 1). The sequences and amino acids of humans and mice are depicted in Fig. 2.
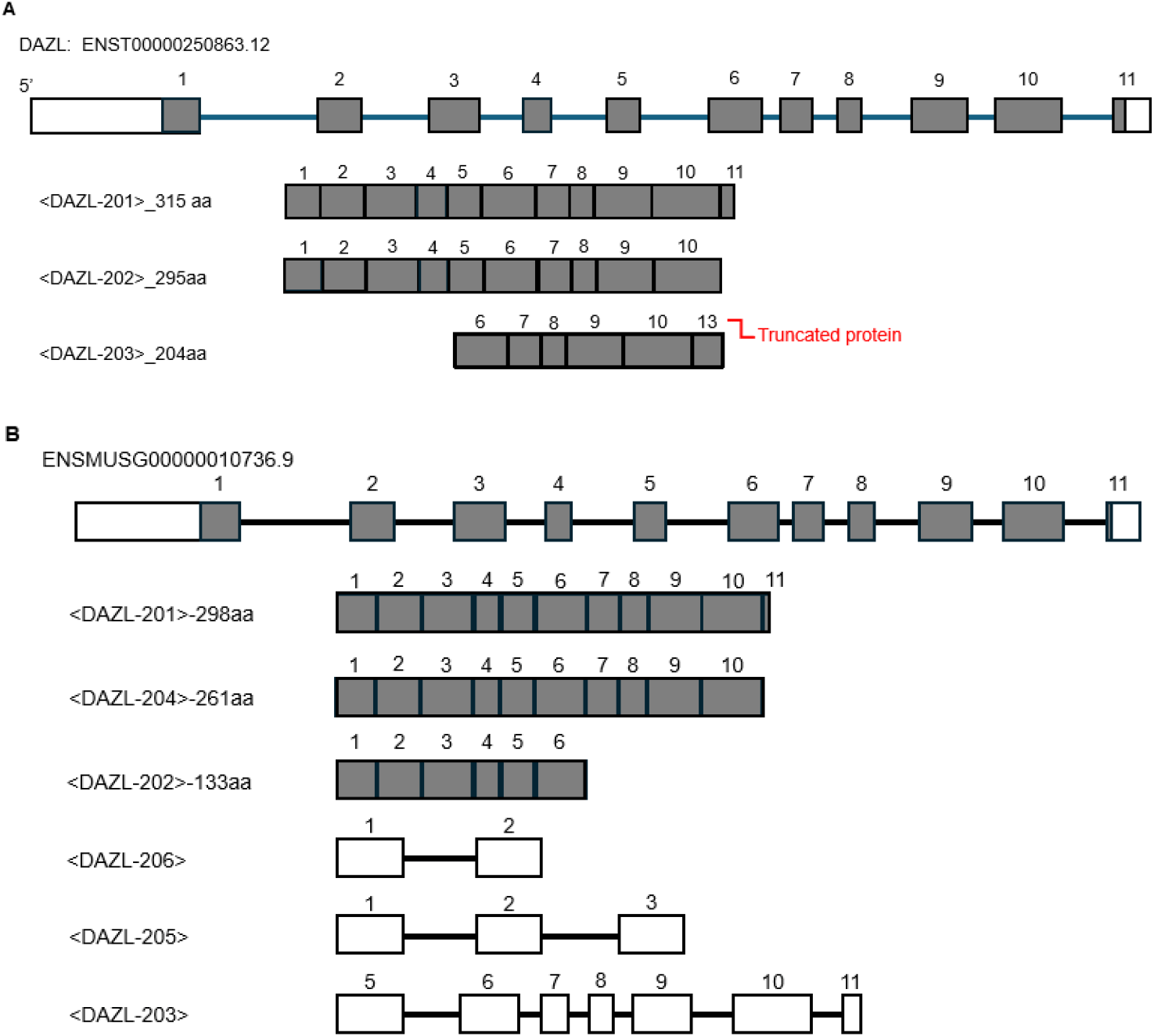
Its expression is detected in several tissues such as the gonad, reproductive system, exocrine system, urinary system, and cleavage stage embryos. It is known that there are 6 splice variants (Dazl-201, 202, 204, 203, 205, and 206) of DAZL proteins in mouse and 3 (DAZL-201, 202, and 203) in human based on alternative splicing (Dyer et al., 2025). Interestingly, in our laboratory, 3 kinds of variants (Dazl-201, 298 aa; azl-202, 133 aa; and Dazl-204, 261 aa) can be identified at the different cell types of decidua at the gestation day 7 mouse uterus (unpublished) using single-cell RNA long-reading sequencing (Fig. 3, Table 1). This suggests that the splice variants may be play roles in decidual differentiation and cellular heterogeneity.
| Gene ID | Ensemble ID | Cell types | |||
|---|---|---|---|---|---|
| S1 | S2 | S3 | Unass | ||
| Dazl 201 | ENSMUSG00000010736 | 0.28 | 0.36 | 0 | 0 |
| Dazl 202 | ENSMUSG00000010592 | 0 | 0 | 0 | 3.72 |
| Dazl 204 | ENSMUSG00000010592 | 0.28 | 0.36 | 2.52 | 0 |
Epigenetic regulation of DAZL may underlie conserved germ-cell-specific expression. The unmethylation of the promoter region, especially the 149 bp of promoter region, harboring several putative Sp1-binding sites, is the cause of activation of the DAZL promoter, and demethylation induces DAZL expression (Linher et al., 2009; Zhang et al., 2016). Post-transcriptional regulation is also an important mechanism. DAZL expression is repressed post-transcriptionally by NANOS2, a a Nanos RNA-binding protein that interacts with DAZL as an antagonist (Kato et al., 2016). In Fig. 2, the sequence for NANOS2 binding sites (Codino et al., 2021) also depicted.
DZA family proteins have been proposed to function as activators of their translation. The DAZL protein typically consists of an average of 271 amino acids (human) and 231 amino acids (mouse) (Dyer et al., 2025). DAZL works either to enhance or suppress translation, depending on tight regulation of the timely production of proteins critical for meiosis, cell cycle progression, germ cell survival, and differentiation (Yan et al., 2022). The cellular localization of DAZL includes the nucleus and cytoplasm. For example, DAZL protein is localized to the nucleus of spermatogonia but relocates to the cytoplasm during meiosis, where it persists in spermatids and spermatozoa.
MOLECULAR STRUCTURE AND MECHANISMS OF DELETED IN AZOOSPERMIA LIKE
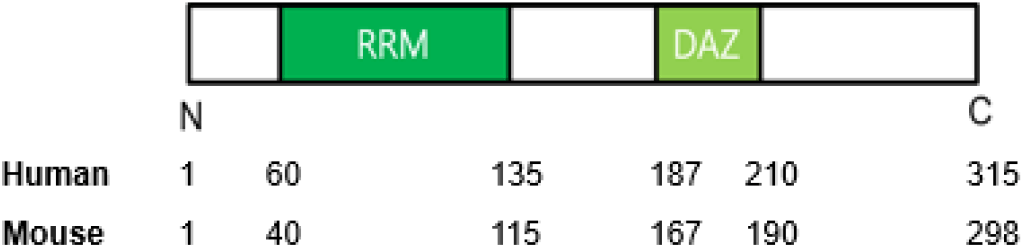
The functional versatility of DAZL as an RNA-binding protein is rooted in its highly conserved molecular architecture. The DAZL protein has two main domains essential for its biological activity: the RNA Recognition Motif (RRM) domain and one DAZ repeat domain (24 amino acids motif rich in Asn, Tyr and Gln) (Fig. 4). In human DAZL, a single DAZ repeat with more closely related to DAZL gene in the mouse (Seboun et al., 1997). The RRM domain works for binding to target mRNAs, while the DAZ domain may work in protein-protein interaction (Tsui et al., 2000). These structural motifs collectively facilitate DAZL’s ability to regulate post-transcriptional gene expression, particularly at the level of mRNA translation (Yan et al., 2022).
RNA RECOGNITION MOTIF (RRM) OF DELETED IN AZOOSPERMIA LIKE REGULATES TRANSLATION
The RRM is the most functionally characterized domain in DAZL and spans about 90 amino acids. The RRM in DAZL recognizes and binds to ‘GUU-’ and ‘UGUU(U/A)-rich’ motifs within the 3′UTR of target mRNAs. Binding specificity has been demonstrated using crosslinking immunoprecipitation (CLIP) and RNA immunoprecipitation-Seq assays reveal that DAZL preferentially associates with transcripts involved in meiosis, chromatin remodeling, spindle assembly, and synaptonemal complex formation. Importantly, mutations or deletions in the RRM impair RNA-binding efficiency and abrogate DAZL function, highlighting its central role in target recognition (Jenkins et al., 2011; Yan et al., 2022). For example, it is reported that DAZL binds to thousands of testicular mRNA transcripts (at least 3,008) at the 3’-UTR (Zagore et al., 2018; Li et al., 2019).
DAZ DOMAIN IN DELETED IN AZOOSPERMIA LIKE REGULATES TRANSLATION
DAZL protein orchestrates a broad translational program that amplifies protein levels of key molecules. For example, DAZL protein orchestrates spermatogenesis and gene regulatory factors to promote the expansion and differentiation of progenitor spermatogonia (Mikedis et al., 2020).
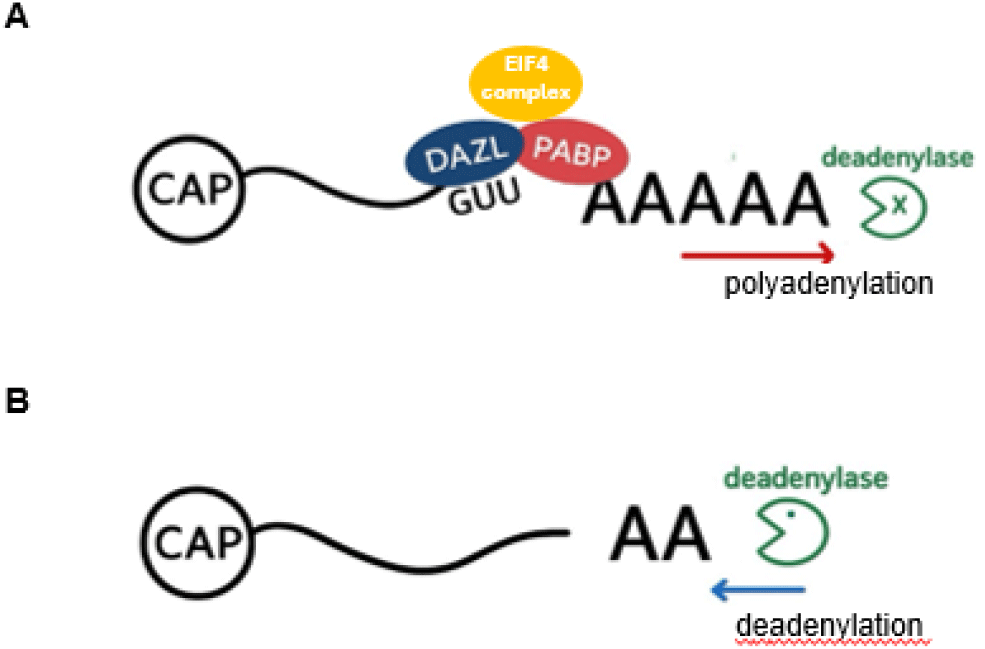
DAZ domain-short conserved peptide sequences mediates interactions with other RNA-binding proteins or components of the translation machinery. This domain may enhance DAZL’s ability to form RNA-protein complexes (ribonucleoproteins, or RNPs), facilitating the recruitment of translational activators such as poly(A)-binding proteins (PABPs), eIF4G, and ribosomal subunits (Fig. 5). Although many germ cell mRNAs possess relatively short poly(A) tails, DAZL facilitates the recruitment of PABPs such as cytoplasmic polyadenylation element binding protein 1 (CPEB1), which in turn promote translation initiation in a poly(A)-tail-length-independent manner (Sousa Martins et al., 2016; Zagore et al., 2018; Li et al., 2019). Panula et al. (2016) showed that DAZL suppresses the translation of OCT4 in embryonic stem cells and affects the transcription of several genes associated with germ cells, cell cycle arrest, and cell migration.
CYTOPLASMIC LOCALIZATION AND STRESS GRANULE FORMATION
In both spermatogenic and oogenic cells, DAZL predominantly localizes to the cytoplasm, often concentrated in ribonucleoprotein granules such as stress granules. These subcellular structures act as hubs for post-transcriptional regulation, including mRNA storage, degradation, and delayed translation. DAZL’s presence in these granules suggests that it may function not only to initiate translation but also to temporally repress or store transcripts stably until they are needed during specific stages of germ cell maturation (Lee et al., 2006; Fu et al., 2015).
DIRECT BINDING TO PRECURSOR miRNAS AND REGULATION OF miRNA BIOGENESIS
The interaction of DAZL and miRNA has been evaluated (Takeda et al., 2009). A recent study (Yan et al., 2022) demonstrated that DAZL binds directly to the loop region of pre-miRNAs containing GUU motifs, particularly members of the let-7 family. This binding enhances DICER-mediated cleavage, significantly increasing mature miRNA levels. This discovery marks the first known role of a germ cell-specific RBP in miRNA biogenesis. Through high-resolution RNA FISH and immunofluorescence on 16-week-old human fetal ovaries, the study demonstrated strong cytoplasmic co-localization of DAZL with let-7a-5p, a member of the let-7 miRNA family, and with DICER, but not co-localize with DGCR8, a nucleus-localized factor in primary miRNA processing. It indicates that DAZL acts specifically during the cytoplasmic DICER-mediated step of miRNA processing.
SPECIFICATION OF DELETED IN AZOOSPERMIA LIKE AND PRE-miRNA INTERACTION
Biochemical assays, including electrophoretic mobility shift assays (EMSA) and fluorescence polarization, revealed that recombinant DAZL directly binds the loop region of pre-miRNAs containing GUU motifs. This loop-specific interaction is sequence-dependent: mutation of the third U in the GUU motif to A significantly reduced binding affinity. Further, truncated versions of DAZL indicated that its RRM domain alone is sufficient for strong binding, while full-length DAZL is required for DICER activation (Yan et al., 2022; Qiao et al., 2025). In summary, the full-length structure of DAZL is required to ensure efficient miRNA cleavage, and its specific interaction with precursor miRNAs influences the processing efficiency, ultimately facilitating the generation of various mature miRNAs.
DELETED IN AZOOSPERMIA LIKE MODULATES PROLIFERATION VIA miRNA IN HUMAN PRIMORDIAL GERM CELLS (hPGCs)
Gene ontology analysis of DAZL-upregulated miRNA targets reveals strong enrichment in proliferation-related pathways. Immunostaining of 12-week-old human fetal ovaries demonstrated that cells with high DAZL expression exhibited significantly lower levels of the proliferation marker KI-67. This effect is mediated through the upregulation of specific miRNAs that inhibit the expression of proliferation-associated genes such as TRIM71. In vitro, DAZL overexpression reduced EdU incorporation rates, indicating diminished DNA synthesis, and led to a shift in cell cycle distribution—specifically, a decrease in 2N (G1) cells and an accumulation of 4N (G2/M) cells. These findings suggest that DAZL limits the proliferation of late-stage primordial germ cells (PGCs) by enhancing the maturation of specific miRNAs, such as members of the let-7 family, and downregulating the expression of proliferation-promoting genes, including TRIM71 (Cheng et al., 2022; Yan et al., 2022).
DELETED IN AZOOSPERMIA LIKE IN GAMETOGENESIS AND EMBRYO DEVELOPMENT
DAZL protein is an essential marker of vertebrate germ cell development. It is suggested that DAZL is necessary to restrict the developmental potential of the germline in various mammals such as mice and humans (Nicholls et al., 2019). In addition, DAZL is required for germ cell competence to respond to retinoic acid, which induces meiosis (Soh et al., 2015). Deletion of the DAZL gene of germ cells at the postnatal stage does not induce female infertility but induces complete male sterility with gradual loss of spermatogonial stem cells, meiotic arrest, and spermatid arrest (Li et al., 2019). The absence of DAZL results in impairment of both expansion and differentiation of the spermatogonial progenitor population. Response to heat is critical in spermatogenesis, and DAZL is involved in the formation of stress granules, and plays a protective role against stress-induced apoptosis (Kim et al., 2012).
DAZL is involved in cyst breakdown through translational regulation of Tex14, and in primordial follicular oocytes, DAZL is translationally suppressed in a 3’-UTR-dependent manner (Fukuda et al., 2018; Rosario et al., 2019). In fully-grown oocytes, the DAZL protein increases 4-fold during reentry into the meiotic cell cycle. Depletion of DAZL causes a block in maturation and causes translational activation of a distinct subset of mRNAs both in quiescent and maturing oocytes, a functional recapitulation with YFP-3’UTR reporter (Yang et al., 2020). DAZL cooperates with CPEB1 to regulate maternal mRNA translation. Ribosome loading is induced in a DAZL- and CPEB1-dependent manner as the oocyte reenters meiosis. The knockdown of DAZL in fully grown mouse oocytes displayed reduced translation activation and improper spindle assembly and fail to get developmental competence (Saunders et al., 2003; Sousa Martins et al., 2016; Yang et al., 2020).
DAZL is also required for normal pre-implantation development. DAZL protein is detected from the zygote stage embryos to the blastocyst (Cauffman et al., 2005; Pan et al., 2008). DAZL knockdown causes a block of the maternal-to-zygotic transition. DAZL morpholinos almost completely block at the two-cell stage, and it is rescued by coinjection of human DAZL mRNA (Chen et al., 2011). In the case of DAZL overexpression mice, the number of pups is fewer than the control due to defective pre-implantation development (Fukuda et al., 2018).
DELETED IN AZOOSPERMIA LIKE IN mRNA TRANSPORT
DAZL protein plays a role in the transport of specific mRNA via the dynein motor complex. DAZL protein functions as an adaptor for a set of mRNA on the dynein-dynactin motor complex and is localized on microtubules. The DAZL-bound mRNAs accumulate in RNA granules at specific sites near the nucleus along the microtubule network (Lee et al., 2006; Smorag et al., 2014).
CANCER-GERMLINE GENE DELETED IN AZOOSPERMIA LIKE
Cancer and germline development share several molecular hallmarks, including sustained proliferation, pluripotency, and mechanisms of immune privilege. These parallels have led to the identification of ‘cancer-germline genes’, a class of genes normally restricted to germline tissues but aberrantly re-expressed in tumors (Xu et al., 2020; Bruggeman et al., 2023). Emerging evidence suggests that DAZL is a member of cancer-germline gene group and plays significant roles in cancer. Its dysregulated expression has been observed in a variety of malignancies such as glioblastoma, non-small cell lung cancer (NSCLC), and testicular germ cell tumors (TGCTs). Importantly, DAZL’s role in cancer appears to be context-dependent-acting as a tumor suppressor in some settings and as an oncogenic driver in others as expected form the roles of DAZL in other cell types (Zhou et al., 2024).
Tewari et al. (2024) explored the DAZL expression in orchiectomy samples from 73 patients with clinical stage 1 TGCTs. In teratocarcinoma models, DAZL overexpression reduces tumorigenic potential, limiting both in vitro proliferation and in vivo tumor formation in xenografted mice. Moreover, its expression correlates inversely with proliferation markers like KI-67, reinforcing its function as a cell cycle brake. These findings highlight DAZL’s potential to constrain aberrant cell growth (Yan et al., 2022).
While DAZL has demonstrated tumor-suppressive properties, mounting evidence suggests that it may also serve as an oncogenic driver in certain somatic malignancies. Zhang et al. (2020) evaluated that DAZL is upregulated in glioblastoma tissues. Knockdown of DAZL in glioblastoma cells led to decreased proliferation, migration, invasion, and stemness, as well as increased sensitivity to chemotherapeutic agents like temozolomide and doxorubicin (DOX). In addition, Zhou et al. (2024) identified DAZL as a novel cancer-germline gene that promotes progression and cisplatin resistance in NSCLC. DAZL enhances the translation of JAK2 and MCM8, leading to increased cell proliferation, viability, migration, invasion, colony formation and DNA repair capabilities. Silencing DAZL suppressed DAZL-mediated cisplatin resistance.
CONCLUSION
The DAZL gene codes for DZAL protein, and its expression is under the control of epigenetic and posttranscriptional modification. The enhancer is localized upstream of the promoter in both humans and mice. DAZL protein has two domains, and they are involved in RNA-binding. Its core functions in regulating mRNA translation, polyadenylation, and microRNA biogenesis position as a central post-transcriptional regulator in normal germline biology. In addition to the possible roles in embryo development, stem cell differentiation, etc., recently it emerged that DAZL is a context-dependent player in tumors. It suppresses tumorigenesis by repressing pluripotency programs and promoting germ cell commitment. In contrast, in somatic malignancies such as glioblastoma and NSCLC, it promotes tumorigenesis by enhancing proliferation, invasion, stemness, and chemoresistance.
On the other hand, the alternative spliced variants of DAZL mRNA are detected, and these can be identified in differentiating tissues and specific cell types. It is suggested that DAZL may involve cell type specificity, although more studies are needed. Collectively, these findings establish DAZL as a gatekeeper of germline integrity, a potential oncogenic factor, and cell type formation. Its roles underscore the complexity of RNA-based regulation in development and disease. Future studies focusing on the tissue-specific interactome and regulatory networks of DAZL will be essential to fully exploit its potential in understanding differentiation during development and as a diagnostic biomarker and therapeutic target in various disorders, including fertility.
