INTRODUCTION
The early stage of development was consisted a complex process including cleavage, blastulation, gastrulation, formation of body axis, tissue and organ (Drasdo & Forgacs, 2000). This complex steps important for successful production of marine fish and increase the survival rate of early larvae. In oviparous animals, accumulation and mobilization of yolk materials are key processes for successful reproduction and the lysosomal system involved in the yolk reservation during embryogenesis for successful development of embryo and early larva. Yolk material accounts for about 80% of the final egg in rainbow trout (Onchorhynchus mykiss) and the embryos are entirely dependent on the yolk material for their growth and nutrition from fertilization to early development (Kwon, 2001). Most oocyte yolk proteins and lipids are derived from the enzymatic cleavage of complex precursors (Wiegand, 1996). Enzymatic cleaved yolk material is stored in the embryo and then provide the nutrients for embryogenesis (Retzek, 1992). Despite the importance of yolk sequestration and mobilization in oviparous animals, the study of this processes is still very limited. Previous several studies suggested that the proteolysis of yolk proteins (YPs) has been implicated as a mechanism for nutrient supply during early development of the embryo in marine teleosts (Carnevali et al., 1998, 1999, 2001). Also, interference of cysteine and aspartic proteinase inhibitors resulted in the blocking of yolk proteolysis and abnormal development or mortality of embryos (Cardinaletti et al., 2010). These results demonstrated that cysteine and aspartic proteinase are involved in the yolk accumulation during oogenesis and its subsequent mobilization in fertilized eggs.
The cysteine proteases including the papain family are a widespread group of proteolytic enzyme that catalyze the hydrolysis of many different proteins and play a major function in intracellular protein degradation and turnover (Bond & Butler, 1987). Cathepsins are widely distributed from microorganism to animal and plant tissues and play a wide range of biological function such as protein turnover, antigen processing, proenzyme activation, hormone maturation and epithermal homeostasis (McGrath, 1999). Especially, cathepsins have been shown to play significant roles in the physiological functions of protein turnover in the lysosome, e.g. antigen processing. These role of cathepsins is applied to antigen-presenting function of the immune system. In general, cathepsin referred to as “lysosomal proteolytic enzymes” and there are composed of cysteine proteases including cathepsins B, C, H, F, K, L, S, W and X (or Z), serine proteases including cathepsins A and G, and aspartyl proteases including cathepsins D and E (Turk et al., 2002). These enzymes shared common protein structures with a signal sequences of 16–18 amino acids, a propeptide of 62–100 residues and then a catalytically active mature region of about 220–230 amino acid (Kirschke et al., 1995) with highly conserved active sites Cys25, His159, and Asn175 (papain numbering) and is folded into a two-domain structure (Brocklehurst et al., 1987). Moreover, the resolution of inflammation depends on apoptosis of inflammatory cells and their subsequent clearance by phagocytes such as macrophages in mammals.
Cathepsin F is most well-studied a member of this enzyme family and has unique extended N-terminal proregion, which contains a cystatin-like domain attached with a flexible linker to the canonical papain-type catalytic domain. In fish, cathepsin F is also known that involved in several important functions such as oocyte maturation (Fabra & Cerdà, 2004), growth of rainbow trout (Kwon et al., 2001) and development of zebrafish retina (Vihtelic et al., 2005).
In previous studies, olive flounder cathepsin F gene has been only reported that molecular cloning, enzymatic characterization and gene expression during immune responses (Ahn et al., 2009; Kim et al., 2010), but the gene expression patterns for early developmental stage after fertilization was unknown. Therefore, the aim of this study was to investigate the gene expression patterns of cathepsin F in various tissues of adult as well as different developmental stages.
MATERIALS AND METHODS
Olive flounders were obtained from Genetics and Breeding Research Center, National Fisheries Research and Development Institute (NFRDI), and maintained in 10 tons flow through tank at 20±1°C under a natural photoperiod. The sample was prepared from various tissues including brain, muscle, liver, intestine, stomach, kidney, skin, fin, spleen, gill, and eye obtained from various aged healthy olive flounder (90-day-old; 10-month-old; 2-year-old; n=3, respectively). Different eight stages of embryo (~0.92 mm) including fertilized egg (F), 1-64 cells (E1), molura (E2), blastula (E3), gastrula (E4), early neurula (E5), mid neurula (E6), late neurula (E7) and six laval stages (~2.49 mm) including 0 to 5 days post hatching (dph) (H, H1, H2, H3, H4, and H5) fish kept at 20°C in the sea-water tank. These samples were observed and determined development process under a microscope imaging systems at ×200, ×300, and ×400 magnification. The sample of 10 randomly selected embryo or fish were collected and immediately frozen in liquid nitrogen, and stored at –80°C until RNA extraction.
Total RNA was extracted using the Trizol Reagent (Invitrogen) according to the manufacturer’s protocol. The total RNA concentration was quantified by spectro-photometer and 1μg of total RNA was used for reverse transcribed into cDNA using First Strand cDNA synthesis kit (Roche). The amplification was performed with AmpliTag Gold DNA Polymerase (Applied Biosystems) in thermal Cycler (Bio-Rad) using the following parameters: denaturation at 95°C for 10 min and 35 cycles of reactions of denaturation at 98°C for 10 sec, annealing at 58°C for 30 sec, and elongation at 72°C for 45 sec. An aliquot of each PCR product was subjected to 1.2% (w/v) agarose gel electrophoresis and visualized by staining with ethidium bromide.
All the primers were designed using Primer3 (Rozen & Skaletsky 2000). The level of cathepsin F expression was detected using specific primers (cathepsin F-F: 5’-GCT CTG AAT GCC TTT GCA AT-3’ and cathepsin F-R: 5’-GTA ACC CTG CTC TCC ATA ATC CT-3’). Housekeeping gene was amplified using beta-actin (ACTB); 5’-GAG CGT GGC TAC TCC TTC AC-3’ and 5’-AGG AAG GAA GGC TGG AAG AG-3’, Genbank accession number (CX725848), forward and reverse primers, respectively.
To evaluate mRNA levels of cathepsin F, these primers were specifically designed to detect and quantify cDNA sequences without detecting genomic DNA. The specificity of the real-time PCR reactions was monitored with melting curve analysis using 7500 software (version 2.0.5). Amplification efficiency was determined by a series of template dilutions. All experiments were repeated in triplicate. Each reactions displayed an efficiency 93.9% (R2=0.993) and 97% (R2=0.999), cathepsin F and ACTB, respectively.
The real-time PCR reaction mixture contained 2 μℓ of diluted sample, PCR primer and the SYBR Green PCR Master Mix (Applied Biosystems). Amplification was carried out as follows: denaturation at 95°C for 10 min and 35 cycles of reactions of denaturation at 95°C for 10 sec, annealing at 58°C for 30 sec, and elongation at 72°C for 45 sec. Thermal cycling and fluorescence detection were conducted using the ABI7500 (Applied Biosystems).
The raw threshold cycle (Ct) represents the PCR cycle at which an increase in reporter fluorescence baseline signal can first be detected. The ratio between the housekeeping gene content in standard samples (106 copies) and test samples were defined as the normalization factor. The fold changes of cathepsin F gene in different developmental stages or tissues were calculated using standard delta-delta-Ct method (Pfaffl, 2001).
Bioinfomatic analysis was conducted to determine gene identities using Gene Master software (Ensoltek, Beaverton, USA). Expressed sequence tags (ESTs) were assembled in clusters of contiguous sequences (conting) using ICA tools program (Parsons, 1995). Gene annotation procedures and homology searches of the sequenced ESTs have been locally done by BLASTX for amino acid similarity comparisons. Matches with the Expect value (E) less than 1.0 × e–4 were considered to be significant. Multiple alignments of the amino acid were constructured using the CLUSTALW program using default parameters (Thompson, 1994) and visualized with MEGA 3.1 (Kumar et al., 2004).
The data are presented as the mean ± S.D. The mRNA levels of cathepsin F were expressed as a ratio to those beta-actin (ACTB), which was simultaneously amplified as an internal control for each cDNA. The data was statistically analized by one-way ANOVA after arcsine transformation when needed, and followed by Duncan’s multiple-range test. All experiments were performed two or three times on different batches and differences were considered statistically significant at P<0.05.
RESULTS
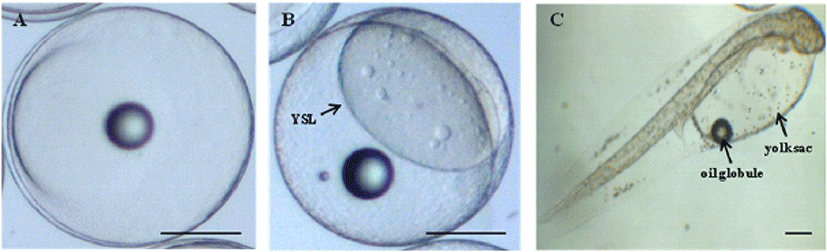
The yolk proteins lead to morphological changes of embryo and lavae in marine fish. Among yolk protein, cathepsin was significantly involved in cellular protein degradation during post mortem, also dead egg was increased in the early developmental stage when the amount of cathepsin decreases. Therefore, in order to monitoring the normal morphological change during early developmental stage in olive flounder, microscopic analysis was performed in embryo and lavae (Fig. 1). The size of fertilized egg was about 0.94 mm after artificial fertilization (Fig. 1A) and yolk syncytial layer (YSL) was clearly observed at higher magnification (Fig 1B) in blastula stage. The size of hatching larvae was about 2.4 mm and has a yolk sac (Fig. 1C). The yolk sac embraces oil globule at its anterior, also has the slender body shape and a pale color by distribution of pigments (Fig. 1C). Abnormal growth and development by various negative factors such as environment, nutrition and development lead to mass mortality and causes adverse effects on next experiment. These result showed that normal size and shape of yolk component observed during morphological change from fertilized egg to hatching egg.
In previous study, we constructed a cDNA library using river samples of olive flounder and we screened 260 clones (unpublished data). The homologs search of the EST sequence with the corresponding amino acid sequence was examined using blast searches of Genbank and sequence alignment was performed with the CLUSTALW program (http://www.ch.embnet.org/software/ClustalW.html). A databank search showed that liver-1-D2 was revealed to be homologous with published flounder cathepsin F (Kim et al., 2010) in olive flounder and has significant high identity above 98%. When compared to cathepsin F of other species were 98% with olive flounder (FJ436098.1), 91% with zebra mbuna (XP_004563245.1), 91% with nile tilapia (XP_003451852.1), 89% with marine medaka (AEA51106.1), 88% with atlantic salmon (ACN10907.1), 88% with ricefish (XP_004073613.1) and 87% with zebrafish (NP_001071036.1) (Fig. 2). Alignment of amino acid residues of the olive flounder cathepsin F gene with those from other fish indicate a high degree of conservation in the amino acid sequence, therefore we suggest that cathepsin of olive flounder will be similar function another cathepsins.

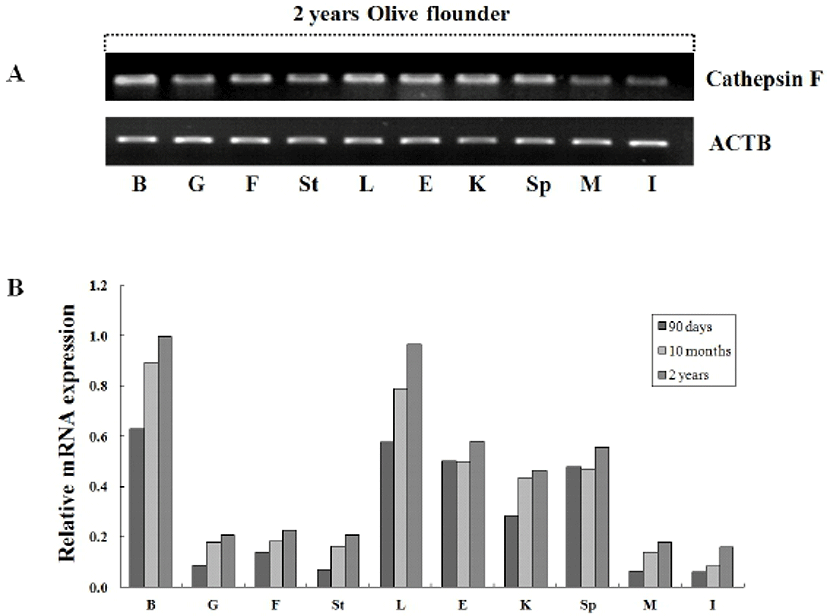
To investigate the tissue specific expression of the cystein protease, oligonucleotide primers were designed according to previously isolated cathepsin F gene in olive flounder (Kim et al., 2010) and then we examined by RT-PCR using isolated various tissues from 2-year-old olive flounder. Transcriptional level of cathepsin was compared between the tissues with beta-actin as the internal gene. Expression of cathepsin F was ubiquitously detected in 2-year-old olive flounder, although somewhat lower expression in muscle and intestine (Fig. 3A). Also, we were analyzed tissue specific expression of cathepsin from various aged olive flounder by real-time PCR. When standardized in the intestine (1.0-fold) of 90-day-old olive flounder, the cathepsin F mRNA level was measured in the brain (10.2-fold), liver (9.4-fold), eye (8.1-fold), spleen (7.7-fold), kidney (4.6-fold), fin (2.3-fold), gill (1.4-fold), stomach (1.2-fold) and muscle (1.0-fold). When normalized to that in intestine of 10 month-old, was in the brain (10.8-fold), liver (9.5-fold), eye (6.0-fold), spleen (5.7-fold), kidney (5.3-fold), fin (2.2-fold), gill (2.2-fold), stomach (2.0-fold) and muscle (1.7-fold). When normalized in intestine of 2-year-old olive flounder, was expressed in the brain (6.3-fold), liver (6.0-fold), eye (3.6-fold), spleen (3.5-fold), kidney (2.9-fold), fin (1.4-fold), gill (1.3-fold), stomach (1.3-fold) and muscle (1.1-fold). Cathepsin was highly expressed in brain, liver, eye, kidney and spleen, while observed five times lower expression in gill, fin, stomach, muscle and intestine in all aged flounder (Fig. 3B). As a result, the mRNA of cathepsin F was ubiquitously and constitutively expressed and dose not has a significant change in tissue specific expression compared with each aged groups. Constitutive expression of cathepsin F in normal olive flounder tissue suggests that it is putative general function as lysosomal protease regardless of pathogen infection or aging.
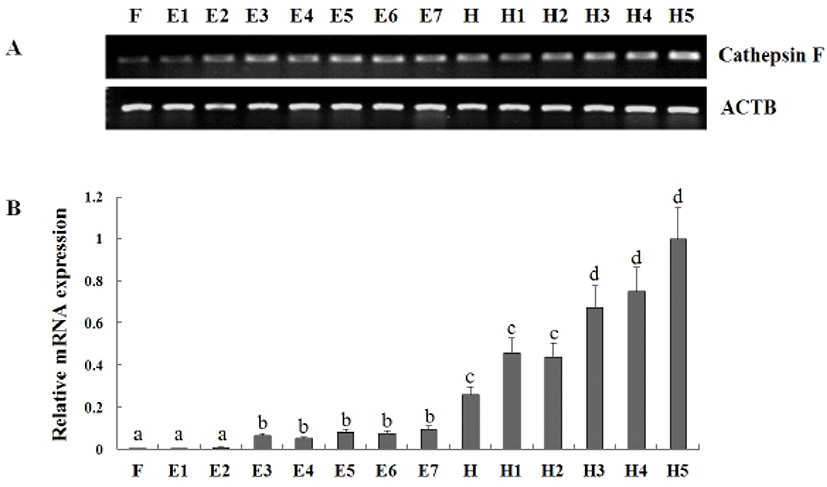
In previous study, Kim et al. (2010) reported cathepsin F that isolated from olive flounder liver cDNA library was evenly distributed all tissues in adult flounder and transcriptional level was gradually increased from 7 dph to 34 dph. But, Fabra & Cerdà (2004) described that cathepsin F mRNA was found throughout the oocyte growth or vitellogenesis period and a transient accumulation during oocyte maturation in teleost Fundulus heteroclitus. To more information of cathepsin F expression during early stage of development, we analyzed the expression pattern of cathepsin F at the eight stages of embryos and six laval stages of days post hatching (Fig. 4). As a result, The mRNA level of cathepsin was already detected from fertilized egg (F) and transcript was gradually increased to 5 days post hatching (dph). Significant induction of cathepsin expression was detected in the blastula, hatching stage, and 3 dph. When compare to that in 1– 64cells stage (E2), the transcript level was 6.9-fold changed in blastula (E3), that in late neurula (E7), the transcript level was 2.72-fold changed in hatching stage (H), and that in 2 dph, the transcript level was 1.53-fold changed in 3 dph. These results showed the mRNA of cathepsin F was accumulated from embryogenesis period to early developmental stage and increased with growth. The temporal expression pattern of cathepsin F during these period suggest a potential role of cathepsin F during embryogenesis-associated process and early developmental stage.
DISCUSSIONS
Because of the importance of lysosomal enzymes during embryo development, many studies have been conducted in fish and these reports commonly indicate that cathepsin genes are involved in yolk protein degradation (Carnevali et al., 2001). Among lysosomal enzymes, cathepsin F cystein proteases may potential role in the yolk processing mechanisms during early developmental stage (Fabra & Cerdà, 2004). Cathepin F was identified in a number of the fish, Paralichthys olivaceus (Olive flounder, FJ436098.1), Maylandia zebra (Zebra Mbuna, XP_004563245.1), Oreochromis nilotic (Nile tilapia, XP_003451852.1), Oryzias melastigma (Marine medaka, AEA51106.1), Salmo salar (Atlantic salmon, ACN10907.1), Oryzias latipes (Ricefish, XP_004073613.1) and Danio rerio (zebrafish, NP_001071036.1) (Fig. 2). The recent study was demonstrated that cathepsin F was ubiquitously expressed in most of the tested tissues by RT-PCR (Ahn et al., 2009; Kim et al., 2010). In this study, to analyze a more accurate patterns of tissue specific expression we examined the tissue distribution of cathepsin F using various tissues from healthy olive flounder of 90 days, 10 month, and 2 year aged. As a result, the expression analysis of cathepsin F shown ubiquitous and constitutive expression regardless of age, with no significant change in expression pattern compared with each aged groups. Significant expressions were mainly observed in lymphoid organs included in liver, kidney, spleen tissues, these expression analysis predicted that cathepsin F will closely relate to immune responses in fish (Ahn et al., 2009; Kim et al., 2010).
According to a recent studies, cathepsin F is speculated that it could be important functions involved in the digestion of the yolk proteins during the embryogenesis. However, the mRNA expression analysis of cathepsin F during embryogenesis and early developmental stages in olive flounder was insufficient. In more detail analysis of cathepsin F expression during embryogenesis and early developmental stage, the mRNA levels of cathepsin F was detected from fertilized egg and gradually increased to 5 days post hatching (dph) and significantly increased in the blastula, hatching stage, and 3 dph. During the blastula stage, extra-embryonic tissue formed can be subdivided into the yolk syncytial layer (YSL) in fish (Carvalho et al., 2010). Cathepsins, cystein proteases are highly expressed in the YSL (Alt et al., 2006), and in other species included in insects (Cho et al., 1999), in sea urchin (Mallya et al., 1992), in zebra fish (Tingaud-Sequeira & Cerdà, 2007) and in salmonids (Kwon et al., 2001; Hiramatsu et al., 2002). Therefore, cystein proteinases, included in cathepsin F have been suggested as the enzymes involved in the processing of yolk materials during early embryogenesis. The high expression of hatching stage (H) and 3 days post hatching (3 dph) was seem to related with degradation process of yolk sac and first feeding. These result may suggest that the proteolysis function of yolk proteins (YPs) has been implicated as a mechanism for nutrient supply during early stages in flounder.
In summary, this study provides the information of cathepsin F mRNA expression during embyogenesis and early developmental stage in olive flounder. The our results show that cathepsin F mRNA level is significantly increased in the blastula, hatching stage and 3 dph which is suggesting that it is important role in yolk processing. However, the more research is needed to the exact function and enzyme mechanism of cathepsin F during developmental stage. Further studies will be necessary to elucidate the specific role of the different embryonic and early lavae including cathepsin F, as well as their interactions and yolk metabolism in olive flounder.

