INTRODUCTION
The innate immune system provides a first line of defense against various microorganisms and is essential for the control of pathogenic infections. The adaptive immune system has evolved to provide a more versatile means of defense which prepares increased protection against subsequent reinfection with pathogen. Although the innate immune system cannot always recognize or eliminate infectious pathogens, play a crucial part in the initiation and subsequent direction of adaptive immune responses, as well as participating in the removal of pathogens that have been targeted by an adaptive immune response. Moreover, because there is delay before the initial adaptive immune response takes effect, the innate immune response has a critical role in controlling infections during this period.
The innate immune system is mediated by pattern recognition receptors (PRRs) that primitive part of the immune system through pathogen-associated molecular patterns (PAMPs) (Magnadóttir, 2006). Lectins belong to the PRR class and a group of sugar binding proteins that recognize the exposed carbohydrates on the cell surface of potential pathogenic microbes, and then agglutinate various cells by binding to cell-surface glycoconjugates (Saraiva et al., 2011). Lectins act as a mediator of self and non-self-recognition and play a significant role in cellular functions, including cell communication, agglutination, proliferation, opsonization, phagocytosis, signal transduction, metastasis and apoptosis (Wassaman et al., 1986; Sharon & Lis, 2004).
A fish lily-type lectin sequence was identified in snakehead murrel (Channa striata), large yellow croaker (Larimichthys crocea), spotnape ponyfish (Leiognathus nuchalis), bartail flathead (Platycephalus indicus), turbot (Scophthalmus maximus), rock bream (Oplegnathus fasciatus), orange-spotted grouper (Epinephelus coioides) (Arasu et al., 2013; Tasumi et al., 2016). In fish, several articles were published that lectins are involved in the immune response against pathogen infection (Choi et al., 2015; Kim et al., 2011; Park et al., 2012; Thulasitha et al., 2016). However, these studies are limited to information that the lectin is involved in the innate immune response.
The rock bream (Oplegnathus fasciatus) is a commercially important marine fish and it has a high economic value in the South Korea. Although the recent rapid development of the rock bream farming, the spread of bacterial and viral diseases causing enormous economic loss of the aquaculture industry. Rock bream iridovirus (RBIV) is one of the major pathogens in rock bream aquaculture and identified as the most important pathogen infecting rock bream in the last decade (Inouye et al., 1992; Nakajima & Sorimachi 1994). However, there is little available information based on temporally integrated observations in the immune responses against virus infection. Therefore, research on immunity-related genes in rock bream is necessary to cope with widespread disease infection.
In the previous, OfLTL-1 has been named as RbLTL and reported that changes of expression in the ontogeny, spatial and pathogen infection response (Park et al., 2016). Although the cDNA sequence of the OfLTL-2 and -3 has been reported, no relevant study on the in vivo function of OfLTLs has been conducted. In this study, we investigated that domain analysis and sequence alignment, expression changes in the developmental stage, tissue-specific distribution and expression profile in the immediate and long-lasting immune response against RBIV infection.
MATERIALS AND METHODS
The cDNA and amino acid sequences of OfLTLs were analyzed using the Basic Local Alignment Search Tool (BLAST) at the National Center for Biotechnology Information website (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and the Expert Protein Analysis System (http://www.expasy.org), as reported previously (Wilkins et al., 1999). Sequence alignment was performed using the ClustalX algorithm (http://www.clustal.org/).
The brood stock of rock bream (5 years old) were reared in the Genetics and Breeding Research Center of the National Institute of Fisheries Science (NIFS, Geoje, Republic of Korea) with aerated seawater and natural photoperiod in a 30-ton tank. When the water temperature was 23.0±1.0℃, photoperiod maintained on a 15h light: 9h dark to induce the maturation of gonads. After spawned naturally, the fertilized eggs in the same clutch were collected by a blotting silk net and incubated with weak aeration and fresh seawater at 23.5±0.5℃. After 25 h, the larvae hatched and were placed in a 15-ton tank at a density of 46 larvae/L. Larvae were fed with the L-type rotifer Brachionus plicatilis from 3 to 20 days after hatching (DAH) and Artemia nauplii from 16 to 30 DAH, and then gradually switched to extruded pellet food beginning at 25 DAH. During the rearing time, the rock bream larvae and juveniles were maintained in the conditions as below: water temperature (24.0±1.0℃) and salinity (32.5±0.5%).
Experimental analysis for tissue distribution including brain, eye, gill, intestine, kidney, liver, skin, spleen and stomach samples were dissected from ten healthy rock breams (total length of approximately 10 cm, 5-6 months old) and immediately frozen in liquid nitrogen, followed by storage in a -80℃ freezer until use. The rock breams were anaesthetized prior to experiments involving tissue collection and pathogen injection, and samples were collected under aseptic conditions. Abnormal and diseased fish were excluded in all of the experiments. All fish were acclimatized to experimental condition for 1 week before processing.
For the RBIV challenge experiment, the rock breams were divided randomly into two groups: a control group and a challenged group. The control and challenged fish were injected with 100 μL of phosphate buffered saline (PBS) or a RBIV suspension (102 TCID50 virus/ fish), respectively (Umasuthan et al., 2013). The temperature to which the experimental fish were subjected was controlled at 20℃ using a re-circulation system, without flow and feeding. Challenged fish collected under aseptic conditions at 0, 3, 6, 12, 24 and 72 hours post-injection and pooled together in equal amounts and frozen in liquid nitrogen. The pooled fish were ground using a homogenizer and subjected to RNA extraction.
Total RNA was extracted from the ground fish with TRI solution (BSK-Bio Co.) as described in the manufacturer's protocol. The total RNA was treated with DNase-I (SigmaAldrich) to remove genomic DNA contamination. The concentration of total RNA was quantified using spectrophotometrically (BioTek, Gen 5.2) and RNA quality was assessed via electrophoresis in 1% agarose gels. cDNA was synthesized with SuPrimeScript RT Premix (2X) (GenNet Bio) using an oligo (dT) primer. To synthesize cDNA, the reverse transcription reaction was conducted as follows: a mixture containing 1 μg of total RNA, the oligo (dT) primer and RNase-free dH2O was held at 65℃ for 5 min, then placed on ice for 5 min. Collect the contents of the tube by brief centrifugation and add 10 μL 2x SuPrimeScript RT Premix, and then incubates at 60 min at 50℃. The final inactivation reaction was carried out for and 10 min at 70℃. Specific primers for rock bream lectins and β-actin were designed using the Primer 3 program (Table 1).
Expression analysis of rock breams lectins was conducted using quantitative real-time PCR with specific primers, and the mRNA levels of β-actin were used as an internal control. Quantitative real-time PCR was conducted using the ABI 7500 Real-time Detection System (Applied Biosystems) according to the manufacturer’s instructions. The final reaction volume contained 10 μL of Fast SYBR Green PCR Master Mix (Applied Biosystems), 100 ng of cDNA, 0.3 μL of each of the forward and reverse primers. The amplification procedure consisted of an initial denaturation step for 20 seconds at 95℃, then 40 cycles of 3 seconds at 95℃ and 30 seconds at 60℃, followed by a final dissociation stage. Dissociation curve analysis of the amplification products was performed after quantitative real-time PCR to confirm the specificity of the PCR products.
The relative expression ratio of the target gene versus the β-actin gene was calculated using the 2−ΔΔCt method (Pfaffl, 2001). All samples were analyzed with three duplicates, and all data are presented in terms of relative mRNA levels, expressed as the mean±SE (n=3). Statistical analyses were performed with SPSS 17.0 software (SPSS Inc.) and the data were subjected to one-way analysis of variance (ANOVA). Differences were considered significant at p<0.05 (*) and extremely significant at p<0.01 (**).
RESULTS
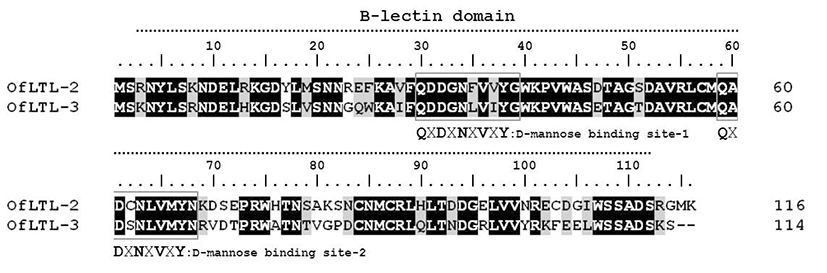
OfLTLs amino acid sequence do not have a signal peptide nor a trans-membrane region, but contains Bulb-type mannose binding lectin(B-lectin) domain between 3 and 112 aa (total of 110 aa sequence) that typical features for its fundamental structure. A long dimerization interface site is also available within the B-lectin domain profile between aa 4 and 112 (total of 109 aa). The deduced amino acid sequence of rock bream lily type lectin-2 shared 69.2 % identity with OfLTL-3. OfLTLs two QXDXNXVXY motifs were completely conserved, but third D-mannose binding domain lack in the rock bream.
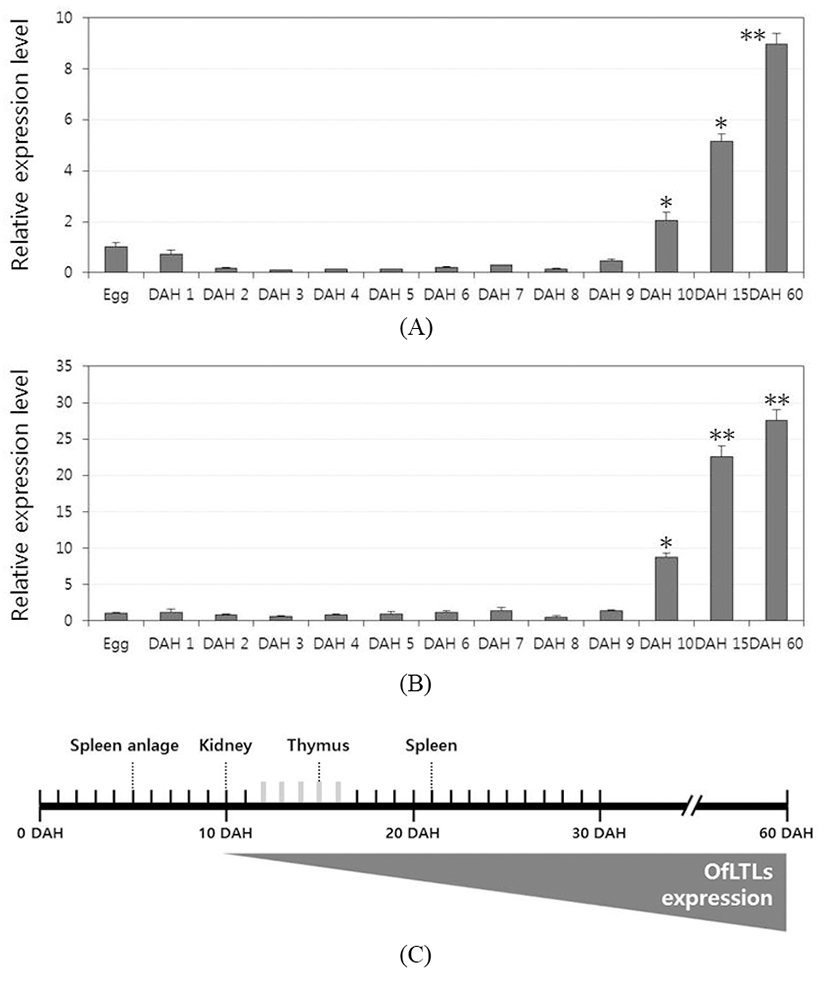
To determine whether a role of rock bream lily-type lectins in the early developmental stage, temporal expression analysis was conducted using the samples from immediately after hatching to 60 days larvae and juveniles rock bream fish. For a quantitative analysis, quantitative real-time PCR was carried out using gene-specific primers designed from the OfLTL-2 and 3 coding sequence. Expression of OfLTL-3 was maintained the basal level without substantially increased until 9 DAH, and it began to increase from 10 DAH and rapidly was increased up to 60 DAH (Fig. 2B). The overall temporal expression pattern of OfLTL-2 is similar to OfLTL-3, although tran sient expression decreased in the early stage that is different with OfLTL-3 (Fig. 2A). In conclusion, OfLTLs are not to expression from the early development, is an increased expression after immune-related organs is fully formed.
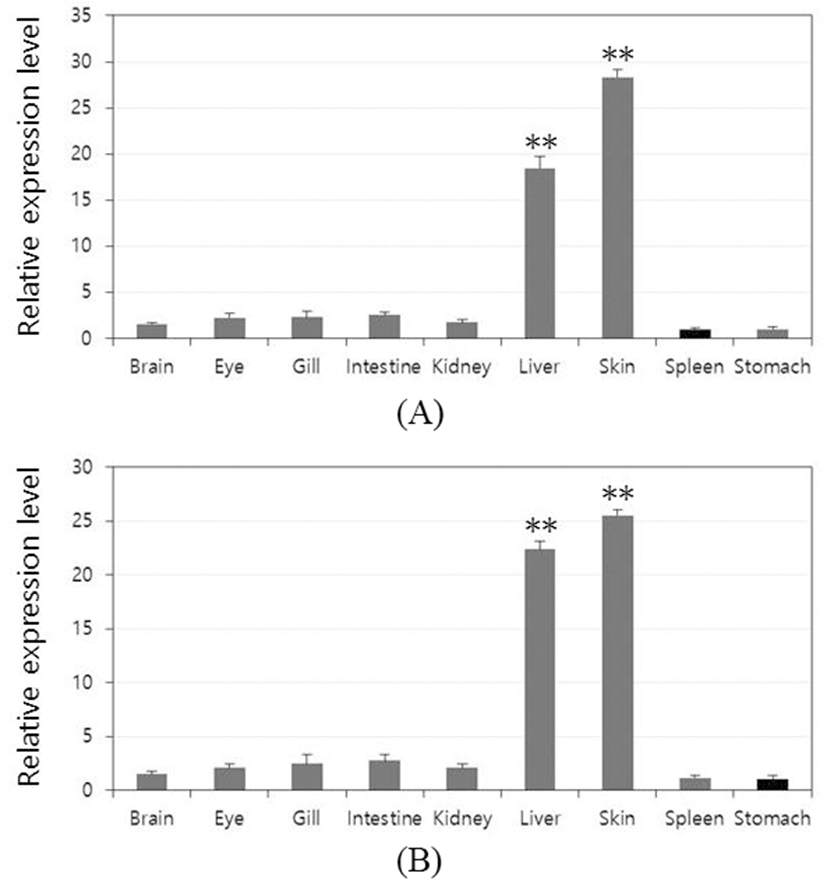
To investigate the role of the rock bream lectins in vivo, it was observed within the tissue distribution. Relative mRNA expression of each tissue was calculated using rock bream β-actin as a reference gene and the result was further compared with the lowest expression tissue to determine the relative tissue-specific expression profile (set as 1). OfLTL2 and 3 were predominantly expressed in the liver and skin that the highest expression in the skin and the following order was liver. Also, the expression of LTLs was almost at basal level except the liver and skin tissue, although the difference between the high-order depending on the gene (Fig. 3A and B).
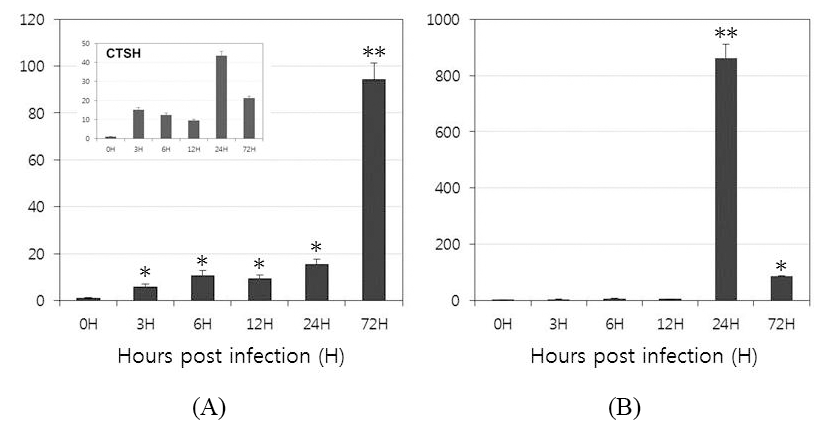
To investigate the function of OfLTLs in the immune response to a viral infection, the temporal patterns of OfLTLs mRNA expression were examined using the whole body of rock bream fish infected with RBIV. Expression patterns and timing are preferentially confirmed with RBIV-induced marker genes to verify a successful challenge experiment. The mRNA level of cathepsin H expression was high in both the early (3H) and later stages (24H); these results are consistent with previous findings (Kim et al., 2013). As shown in Fig. 4, although expression of OfLTL-2 was weak in the early response period, it gradually increased until 24 hours post-infection and then dramatically increased to 72 hours. While, expression of OfLTL-3 maintaining almost the basal levels until 12 hours post-infection and it was exponentially in infected 24 hours post-infection, and then increased expression of OfLTL-3 was reduced to 1/9 at 72 hours after infection.
DISCUSSION
In contrast to higher vertebrates, fish is living in aquatic environments from early embryonic stages of life, which are an ideal medium for microorganism growth compared to air and every moment is constantly exposed to a pathogen attack. The bony fishes are derived from one of the earliest divergent vertebrate lineages to have both innate and acquired immune systems. However, the adaptive immune response did not clearly reveal in fish and it is expected not sophisticated like mammals. Therefore, the innate immune system is the only defense weapons of invertebrates and a fundamental defense mechanism of fish and more dependent their innate immune systems for survival and the resistance to the pathogens.
Lectins play important roles in the recognition and elimination of pathogens via the innate immune system. Researches on the immunological functions of lectin are concentrated in the innate immune response of higher vertebrates. However, our results seem to be involved both in the innate and adaptive immune response; lily-type lectins are expressed after completing immune system during early developmental stage and preferentially increased in the second half reaction in a viral challenge, these results are related to the adaptive immune response. On the other side, OfLTLs are expressed in the innate immune response related tissue. Because fish have not sophisticated and specialized the adaptive immune systems, the innate immunity-related genes will be able to engage in the long-lasting immune responses to infection. Research findings are not infrequently has been reported that the early and late results are quite cross. Therefore, outside the framework defined in the higher vertebrates various studies will be needed.
In the development of the immune system rock bream, spleen anlage was observed between the swim bladder and the intestine at 5 DAH, and the thymus was formed as a paired structure under the pharyngeal epithelium above the gill arch at 10 DAH. The order of the immune organs becoming lymphoid was the pronephric kidney (10 DAH), thymus (15 DAH) and spleen (21 DAH) (Zhizhong et al., 2013). OfLTLs were hardly expressed in the initial developmental phase and increased from 10 DAH. Thereafter, expression of OfLTLs was proportionate to development of rock bream immune organ. It can be inferred that they will participate in the immune response after the immune system is fully established, not involved from the initial development period.
Fish are permanently exposed to various external hazards that aerobic and anaerobic bacteria, viruses, parasites, marine pollutants. To protect fish from pathogenic microorganisms, mucus acts as a physical, chemical and biological barrier between the fish and the environment which first site of interaction with pathogens. The mucus composition is very complex and includes immuneglobulins, agglutinins, lectins, lysins and lysozymes that are anti-microbial factor and is secreted from the fish skin. These factors are very important recognize the microorganisms invasion and to protect the fish from invading pathogens (Lee et al., 2015; Suzukiet al., 2003.
The liver is unique anatomical and immunological organ: The typical function of the liver is carbohydrate, protein and lipid metabolism, bile secretion, antioxidation and detoxification. As well as, the liver play a critical roles against invading pathogen in the innate immune responses of fish that particularly enriched macrophages and lymphocytes compared with other organs and modulated of liver injury and recruitment of circulating lymphocytes (Racanelli & Rehermann, 2006; Castro et al., 2014).
Although it does not clearly separated innate and acquired immune response dedicated organ in fish, liver and skin are primarily involved in many innate immune response. In this study, OfLTLs are predominantly expressed in the liver and skin. These results are to be expected that OfLTLs contribute to the innate immune response against viral infection, but the transcripts were increased to more intensively in the period of acquired immune of RBIV challenge experiments. For this reason, the further expression analysis is needed using a virus-infected liver and skin tissue of the fish because it uses the RSV infection whole fish in this experiment.
