INTRODUCTION
Before the application of CRISPR/Cas9(clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9) technology in the zebrafish research field, targeted gene knockouts were not possible in zebrafish, and its utility for validation studies of candidate genes was limited. At that time, researchers using the zebrafish animal model have been experienced difficulty using ENU technology to generate mutants for interesting genes. This challenge was eliminated with the development of novel gene targeting approaches including ZFNs, TALENs, and CRISPR/Cas9 (Bedell et al., 2012; Jinek et al., 2012; Mali et al., 2013; Hsu et al., 2014; Varshney et al., 2015). Among these, the CRISPR/Cas9 approach now offers an efficient method to target any gene of interest. Although off-target effects of CRISPR/Cas9 technology remains a hurdle for consideration, several screens and engineering methods have been developed to reduce genome-wide off-target mutations including nuclease mutation, protospacer adjacent motif (PAM) sequence modification, single guide RNA (sgRNA) truncation and novel nuclease discovery. For example, Kleinstiver et al. (2015, 2016) recently reported that manipulation of the Cas9 protein (i.e., SpCas9) and sgRNA target not only improved specificity but also reduced off-target effects (Fu et al., 2014).
Matrix metallopeptidases, also known as matrix metalloproteinases (MMPs), are metalloproteinases that are calcium-dependent, zinc-containing endopeptidases. These enzymes are capable of degrading all kinds of extracellular matrix (ECM) proteins, but can also process numerous bioactive molecules. The most common categorizations of MMP are partly based on the historical assessment of substrate specificity and cellular localization of MMP, which include collagenases, gelatinases, stromelysins, and membrane-type MMPs (MT-MMPs). Among them, MT-MMPs constitute a growing subclass of recently identified MMPs; however, MT-MMPs are yet to be fully understood. Previously, Choi et al. (2017) reported that mmp15b was highly expressed in the regenerating liver after severe hepatocyte ablation. In addition, MMPs are involved in the degree of initial injury and repair, the onset and resolution of inflammation, the activation and deactivation of myofibroblasts, and the deposition and breakdown of ECM. In other words, MMPs are involved in both augmenting and attenuating many processes that impact fibrosis (Giannandrea & Parks, 2014). Thereby, a zebrafish mutant model of mmp15b would be an effective tool to investigate the underlying mechanism during liver fibrosis after severe hepatocyte injury.
In this study, we generated a zebrafish mmp15b mutant using CRISPR/Cas9 technology as a model to explore liver disease with human fibrosis.
MATERIALS AND METHODS
We used wild type (TÜ and AB) zebrafish. Wild-type zebrafish were obtained from the Zebrafish Center for Disease Modeling (ZCDM). Embryos and adult fish were raised and maintained under standard laboratory conditions (Westerfield, 2000).
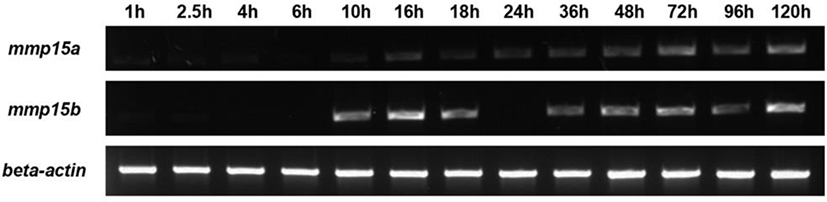
cDNA from embryos at various stages (as indicated in Fig. 1) was used as a template for PCR to amplify mmp15a and mmp15b genes. The primer sequences, mmp15a (407 bp) and mmp15b (556 bp), used for RT-PCR were 5’-GACTTCATGGGATGCCGAGT-3’, 5’-ACGACAAGGTGTGTGTCTCG-3’, and 5’-CCGGTTACCCTCAAGAGCTG3’, 5’-GTGCGCTCCGTCTCATCTAT-3’; respectively.
mmp15b sgRNAs (5’-ATGACCCGTTCACGTTGCTGTGG-3’ and 5’-TTGATGGGTGCGCGCACCTTTGG-3’) were generated as previously described (Irion et al., 2014). mmp15b target sequences are located in exon 1. mmp15b gRNAs (500 ng each) and Cas9 protein (1 ug) were mixed and injected into 1-cell stage embryos which were raised to adulthood. Cas9 protein was purchased from Toolgen. Either the adult zebrafish tail fin or the whole embryo at 1–2 dpf was used to obtain genomic DNA for PCR-mediated genotyping. PCR products were sequenced to identify frameshift mutation. F1 fish containing a 14 bp deletion were selected to establish the mmp15b mutant line.
Results and Discussion
We recently established a zebrafish liver regeneration model in which severe hepatocyte loss resulted in the regeneration of hepatocytes from BECs. Using this model, we performed RNA-Seq analysis to identify pathways or factors involved in the regulation of liver regeneration (Choi et al., 2017). Among them, we selected mmp15b which was highly expressed in early liver regeneration. MMPs are a family of extracellular endopeptidases defined by conserved catalytic domains (Ra & Parks, 2007) and have long been considered to be primarily responsible for turnover and degradation of ECM substrates. However, they are now recognized for immunity and repair; being involved in such processes as cell migration, leukocyte activation, antimicrobial defense, chemokine processing and more (Yoshifum, 2015). Thereby, mmp15b might play a regulatory role in zebrafish liver regeneration or liver fibrosis.
Due to genome duplication in zebrafish, we found that two mmp15 genes, mmp15a and mmp15b, searched. We first examined the expression pattern of mmp15 genes during development. Expression of mmp15a and mmp15b was detected as early as 10 hpf (hour post-fertilization), indicating zygotic but not maternal expression (Fig. 1). Based on their expression patterns, mmp15a and mmp15b likely play an important role in normal animal development and in other physiological settings. Of note, gene expression profiling after RNA-Seq analysis during early liver regeneration showed that mmp15b was highly induced in the regenerating liver, whereas mmp15a was not (Choi et al., 2017). These results suggest a role of mmp15b during liver regeneration.
We generated mmp15b zebrafish mutant utilizing CRISPR/Cas9 technology which is a simple yet powerful tool for editing genomes by altering DNA sequences and modifying gene function. We first designed sgRNAs (Fig. 2B) after predicting the critical domain of Mmp15b (Fig. 2A). As an MT-MMP, the Mmp15b protein contains a membrane linker, cytoplasmic tail, and catalytic domain for the degradation of ECM substrates (Fig. 2A). Therefore, we synthesized two sgRNAs in exon 1 of mmp15b (Fig. 2B) and tested the efficiency by co-injecting with Cas9 protein into one-cell stage embryos. After performing T7E1 assay for DNA sequence mismatch in co-injected embryos, we found that the sgRNAs efficiently generated mismatched DNA, suggesting successful generation of the mmp15b mutant zebrafish (Fig. 2C).
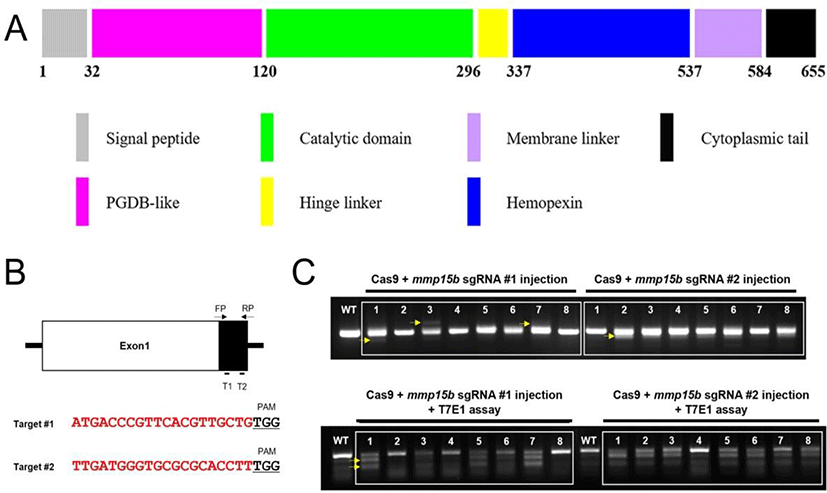
The resulting mismatched DNA from the two sgRNAs suggested germline transmission and were used to generate the mmp15b knockout mutation (Fig. 2C). To determine the role of Mmp15b, we examined (1) outcross with wild type raised to adulthood and (2) identification of mutation by PCR and DNA sequencing in the F1 zebrafish (Fig. 3). We performed PCR after isolation of genomic DNA from 42 zebrafish offspring (Fig. 3B bottom). Approximately 52% (22/42) of zebrafish offspring were mmp15b knockout mutants showing 13, 14, or 16 bp deletions, suggesting that frameshift occurred in the 22 F1 zebrafish (Fig. 3B). Frameshift mutation of mmp15b resulted in termination-site change and early termination of the Mmp15b protein, indicating that the Mmp15b variant was generated with 32 amino acid of the Mmp15b protein. Of note, the mmp15 mutant exhibited no obvious morphological phenotypes in the developing embryo or adult (data not shown). Despite the absence morphological changes, the mmp15b mutant would be a useful tool for the study of liver regeneration. The zebrafish liver is fully functional by day 5, which provides us an opportunity to investigate liver regeneration by combining other transgenic fishes; namely hepatocyte ablation lines, e.g., fabo10a:CFP-NTR, or macrophage ablation lines, such as mpeg1:NTR-mCherry. As our mmp15b mutant zebrafish could be utilized in this regard, future experiments would be of interest to explore the role of mmp15b underlying fibrosis mechanism during liver regeneration.

