INTRODUCTION
Adult stem cells are present in most tissues and provide differentiated cells when needed throughout an animal’s entire life (Kretzschmar & Clevers, 2017). This ability is realized through a stem cell’s unique capability for self-renewing cell division, in which it divides to simultaneously generate a daughter stem cell and a daughter cell that initiates differentiation (Funk et al., 2020). This unique property of stem cells is provided by the niche, a local environment that houses stem cells: a daughter cell within the niche’s area of influence maintains stemness and does not undergo differentiation, while one outside the niche loses stemness and starts differentiation (Spradling et al., 2008; Losick et al., 2011). Accordingly, stem cells are attached to niche cells and influenced by signaling molecules from them.
The Drosophila testis offers an incisive genetic animal model for dissection of molecular mechanisms underlying niche specification, aging, and niche-stem cell interaction (Herrera & Bach, 2019). An assembly of ~12 cells known as hub cells is located at the terminal tip of the testis and provides the niche for both germline stem cells (GSCs) and cyst stem cells (CySCs) (Yamashita et al., 2005). Hub cells are directly attached to GSCs and CySCs via adherens junctions, and express and secrete signaling molecules that stimulate GSCs and CySCs to promote stemness, cell division, and their attachment to hub cells (Kiger et al., 2001; Tulina & Matunis, 2001; Shivdasani & Ingham, 2003; Kawase et al., 2004; Leatherman & DiNardo, 2010; Michel et al., 2011).
Within the Drosophila testis, the RNA binding protein Lin28 is exclusively expressed in hub cells (Sreejith et al., 2019). Lin28 not only acts as a repressor of Let-7 biogenesis but also controls the stability and translation of target mRNAs including Unpaired (Upd), an extrinsic stem cell self-renewal factor that is expressed and secreted from hub cells (Kiger et al., 2001; Tulina & Matunis, 2001; Sreejith et al., 2019). A second RNA binding protein from a conserved protein family, IGF-II messenger RNA binding protein (Imp), is also required for stabilizing Upd transcripts (Toledano et al., 2012). However, what other factors Lin28 and Imp may act to stabilize in hub cells is yet largely unknown.
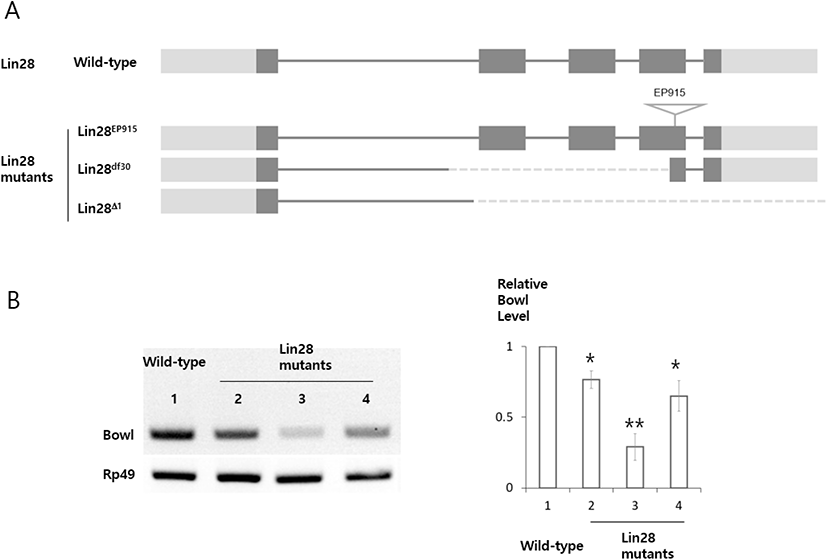
Hub cells and CySCs are both specified during embryogenesis from somatic gonadal precursors (SGPs) (D Le Bras & Van Doren, 2006; Kitadate & Kobayashi, 2010; iNardo et al., 2011; Okegbe & DiNardo, 2011). For hub cells, specification, aggregation, and assembly are known to require Notch signaling and the transcription factor Bowl, while CySC specification requires Lines (DiNardo et al., 2011; Okegbe & DiNardo, 2011). Notably, the embryonic gonads of Bowl mutants contain fewer hub cells, while increased Bowl activity transforms CySCs to the hub cell fate (DiNardo et al., 2011); these findings indicate that Bowl is a hub cell specification factor. Similarly, Lin28 mutant embryonic gonads contain fewer hub cells (Sreejith et al., 2019), suggesting that Lin28 could function in hub cell specification together with Bowl. Here we find that in hub cells, Bowl transcripts require Lin28 and Imp for stability.
RESULTS AND DISCUSSION
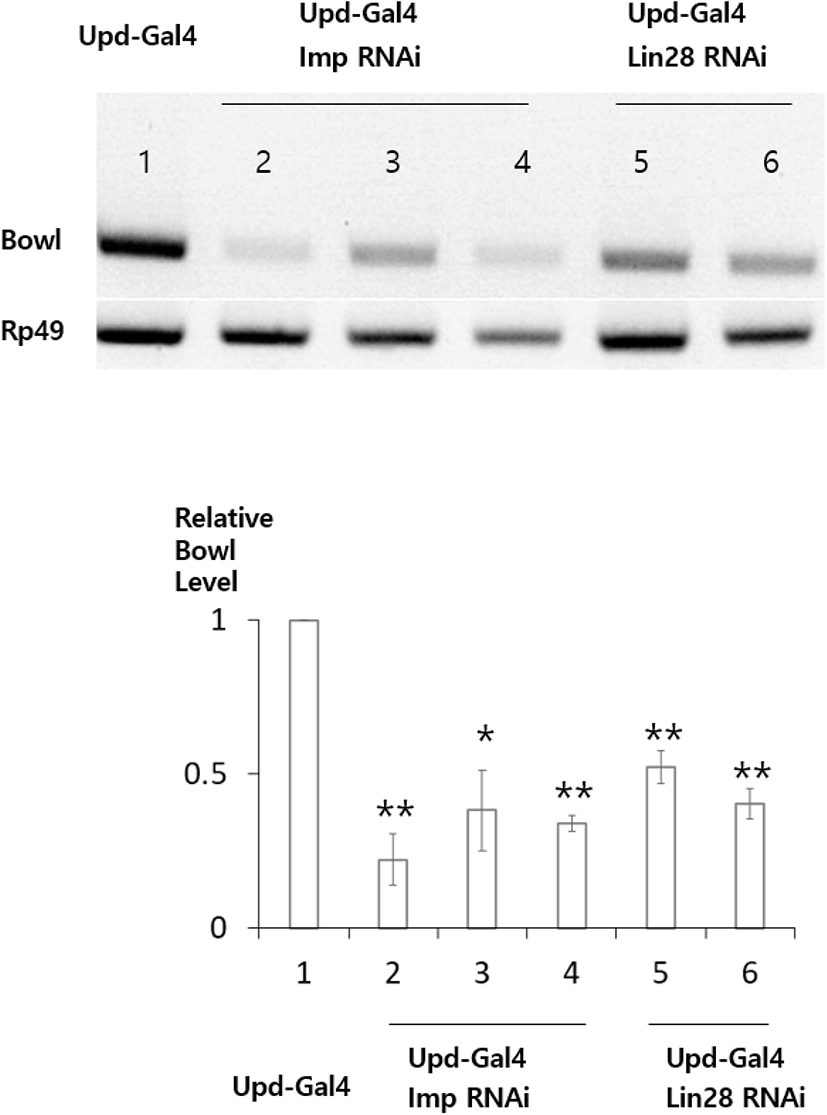
Lin28 was previously shown to be required for stabilizing transcripts of Upd, an important hub cell factor (Sreejith et al., 2019). This observation prompted us to identify other targets that are stabilized by Lin28 in hub cells of the Drosophila testis. To identify new target transcripts, we carried out reverse transcription polymerase chain reaction (RT-PCR) and compared transcript levels in Lin28 mutant and wild-type testis. This screening identified Bowl transcripts as reduced in the Lin28 mutant (Fig. 1). Lin28 is exclusively expressed in hub cells, suggesting that Bowl reduction is due to Lin28 mutation specifically in hub cells. To confirm this notion with an independent method, we used the bipartite GAL4-UAS system (Brand et al., 1994) in combination with RNA-mediated interference (RNAi) to reduce Lin28 function exclusively in hub cells. Transgenic flies bearing Upd-Gal4 (hub cell-specific Gal4) and UAS-Lin28 RNAi were generated (Upd-Gal4>UAS-Lin28 RNAi), in which Lin28 RNAi was specifically expressed in hub cells. RT-PCR of testis samples from two independent Lin28 RNAi lines confirmed the reduction of Bowl transcripts (Fig. 2). We likewise examined the relation of Bowl and Imp; however, Imp null mutation was lethal, so we instead relied on Imp knockdown in hub cells of the adult testis. Testis from three independent lines of Upd-Gal4>UAS-Imp RNAi flies expressing Imp RNAi in hub cells likewise showed reduction of Bowl transcripts (Fig. 2). Taken together, these data suggest that maintenance of Bowl transcripts in hub cells requires both Lin28 and Imp.
To further investigate the molecular mechanisms underlying promotion of Bowl transcript stability by Lin28 and Imp, we carried out cell culture experiments using Luciferase (Luc) constructs bearing the Bowl 3’UTR (Luc-Bowl 3’UTR), employing RT-PCR to examine whether those hybrid transcripts were stabilized by Lin28 and Imp, which stabilizes target mRNAs through 3’UTR (Toledano et al., 2012; Lee et al., 2019; Sreejith et al., 2019). When either Lin28 or Imp was co-transfected with the construct, Luc-Bowl 3’UTR transcript levels were increased (Fig. 3A). Meanwhile, levels of control Luc transcripts lacking the Bowl 3’UTR were not affected by the presence of either Lin28 or Imp (Fig. 3B). These results suggest that Lin28 and Imp stabilize Luc-Bowl 3’UTR transcripts through the Bowl 3’UTR.
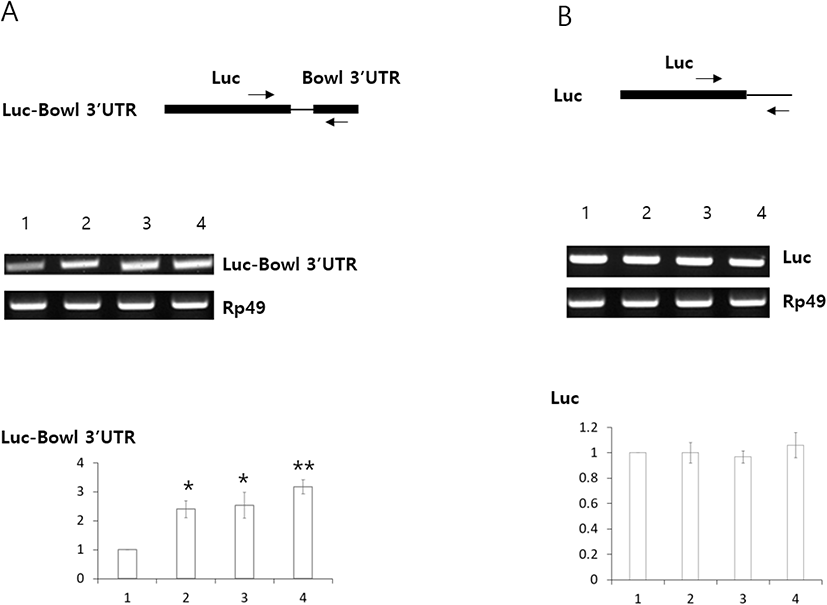
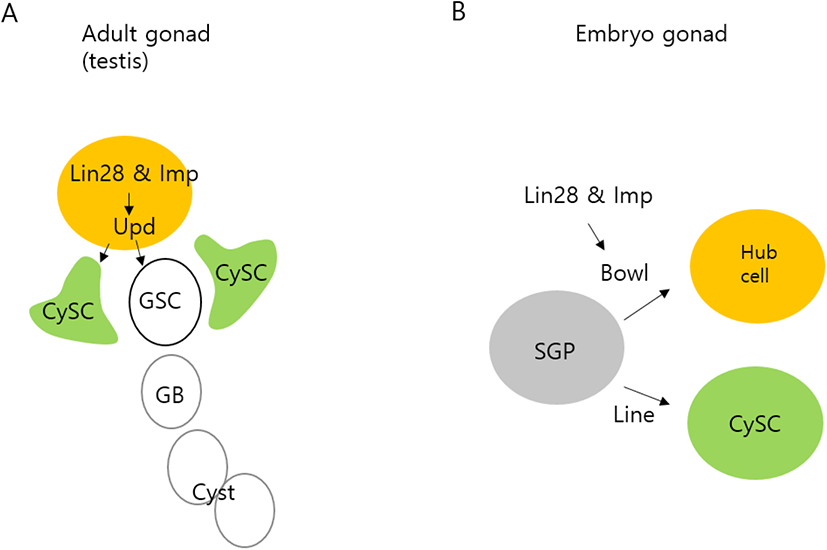
Fig. 4 illustrates our proposed model of the roles of Lin28 and Imp in the testis. We and others previously showed that in adult testis hub cells, Lin28 and Imp are required to sustain hub cell maintenance and function and act through stabilizing mRNAs encoding Upd, a critical hub cell factor that stimulates stemness and cell division of GSCs and CySCs (Fig. 4A) (Toledano et al., 2012; Sreejith et al., 2019). In this communication, we identify a new hub cell-specific mRNA stabilized by Lin 28 and Imp, the Bowl transcript (Fig. 4B). Bowl was previously shown to be a hub cell specification factor during early development of the embryonic gonad (DiNardo et al., 2011). Our findings suggest that Lin28 and Imp are present in hub cells and act to protect both Bowl transcripts in embryos and Upd transcripts in adults. An interesting question remains regarding the mechanistic particulars of how Lin28 and Imp stabilize Upd and Bowl mRNAs. Future studies are required to address this question.
MATERIALS AND METHODS
Flies were grown in standard fly food. W1118, Lin28EP915, and ImpMI05901 were from the Bloomington Drosophila Stock Center (BDSC). Lin28df30 was from F. Michon and Lin28∆1 from N. Sokol. UAS-Lin28 RNAi (50679, 29564) and UAS-Imp RNAi (34977) were from the BDSC and UAS-Imp RNAi (20321, 20322) from the Vienna Drosophila RNAi Center (VDRC).
RNA was extracted using standard TRIzol (Invitrogen) procedure. Complementary DNA synthesis was performed with oligodT primers using one microgram RNA. PCR was performed with the following primers: Rp49: Forward 5’-CACCAGGAACTTCTTGAATCCGG-3’, Re v erse 5’-A GA T C G T GAA GAA GC GC A C C-3’; Bo wl: F or war d 5’-CTGCTCATCCACGAGAGG-3’, Reverse 5’-TGTGGACAGCCAAGGTTC-3’; Luc-Bowl 3’UTR: Forward 5’-CC CTCGAG CAATCCATTAATGGAG-3’, Reverse 5’-GC TCTAGA CATTATTAATGCATACTTTATTTGA-3’. RT-PCR were carried out three replicates in Fig. 1, 2, and 3.
pAc5.1A-Luc (S2 cell expression vector) was created using the Luc gene from the pGL3-Basic vector (Promega, Madison, WI, USA), which was ligated into the EcoRI-NotI sites of pAc5.1/V5-HisA (Invitrogen). The Bowl 3’UTR (853 bp) from Bowl cDNA (RE05342, Drosophila Genetic Resource Centre) was ligated into the XhoI-XbaI sites of the pAc5.1A-Luc vector.
Schneider 2 cells were grown and maintained in Shields and Sang M3 insect Media (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% insect media supplement (Sigma-Aldrich) with appropriate antibiotics at 25 °C. Cells were subcultured every five days at a 1:5 ratio when confluency reached around 100%. The DDAB method of transfection was employed with some modifications for 24-well plates (Han, 1996). Cells were collected and assayed three days after transfection.
