INTRODUCTION
During embryonic head development, all insects build segments that fuse to each other to form the head capsule bearing appendages with different roles in feeding and feeling (Snodgrass, 1935). The insect head consists of the anterior pregnathal and the posterior gnathal regions. While the anterior pregnathal region comprises the intercalary (IC), antennal (AN), ocular segments (OC), and other preantennal tissues, the posterior gnathal region is composed of the mandibular (MD), maxillary (MX), and labial (LB) segments that constitute the mouthparts (Snodgrass, 1935; Rogers and Kaufman, 1997). The genetic mechanisms of head segmentation have been studied in most detail in Drosophilamelanogaster. In Drosophila, the typical hierarchyofmaternal, gap, pair-rule, and segment polarity gene interactions form the maxillary and labial gnathal segments (St Johnston & Nusslein-Volhard, 1992).For patterning the pregnathal and the mandibular segments, however, Drosophila utilizes the head gap genes, orthodenticle (otd), empty spiracles (ems) and buttonhead (btd), which are expressed in broad and overlapping domains of the embryonic head region (Dalton et al., 1989; Cohen & Jurgens, 1990; Finkelstein & Perrimon, 1990; Walldorf & Gehring, 1992; Wimmer et al., 1993). Consistently, mutations in any head gap gene display phenotypes missingthe pregnathal and the mandibular segments that correspond to the expression domains of each head gap gene. For example, otd mutants do not bear the antennal and ocular segments, and ems mutants do not have the antennal, intercalary, and the anterior portion of the mandibular segments (Dalton et al., 1989; Cohen & Jurgens, 1990; Walldorf & Gehring, 1992). In addition, loss of btd leads to the lack of antennal, intercalary, mandibular and the anterior portion of the maxillary segments (Wimmer et al., 1996).
It has been suggested that the genetic mechanisms of head segmentation in Drosophila might not be always applicable to other insect head segmentation. In Drosophila, activation of the head gap genes depends on Bicoid, which is the key maternal factor for patterning anterior regions along the anterior-to-posterior axis of Drosophila embryo (Finkelstein & Perrimon, 1990; Wimmer et al., 1995; Gao & Finkelstein, 1998). Interestingly, however, it has become clear that bicoid is absent from most other insects but probably arose recently in the lineage of dipterans during insect evolution (Stauber et al., 1999; Schroder, 2003; Chouard, 2008). Furthermore, Drosophila larva has the derived head morphology including the maggot's head being hidden within the thorax and the head structures being highly derived (Rogers & Kaufman, 1996). To understand the evolution of genetic mechanisms controlling head development, orthologs of Drosophila head gap genes have been analyzed in insects (Schinko et al., 2008). In the red flour beetle Tribolium castaneum, functional analyses of orthologs of Drosophilahead gap genes revealed that the head segmentation mechanisms in Triboliumare distinct from those of Drosophila.Similar to Drosophila, Tc-otd1 plays as a head gap gene for the early regionalization of all head segments formed during the blastoderm stage and then for the later head patterning (Schinko et al., 2008). However, it has been suggested that Tc-ems and Tc-btd are not head gap genes; Tc-ems has a minor role for patterning the antennal segment and Tc-btd is not required for head development at all (Schinko et al., 2008).The dispensable role for Tc-btd in Tribolium head development is unexpected and remains arguable because Tc-btd is expressed in the future mandibular segment at the blastoderm stage, with its later expression in the antennal and intercalary segments (Schinko et al., 2008). In the milkweed bug Oncopeltus fasciatus, knock-down of Of-otd leads to the lack or reduction of the anterior region to the mandibular segment, with the mandibular and maxillary segments being malformed (Birkan et al., 2011). However, roles for Of-ems could not be analyzed because of the very low penetrance of RNAi effects (Birkan et al., 2011).
Here we reexaminedthe expression patterns and functions of Tc-btd during Triboliumsegmentation for better understanding the role of Tc-btdin Tribolium development.Tc-btdis expressed in stripes with single segmental periodicity at the segmentation stages, with its later expression in the head segments and head lobes. Larval cuticles of Tc-btd RNAi (Tc-btdRNAi) embryos display the loss of pregnathal regions with the gnathal maxillary and labial segments remaining in the head and the defects in abdominal segmentation in the trunk. The defects are observed in the head and trunk regions of the growing germbands undergoing embryonic segmentation. Our observations suggest that Tc-btd is required for the head and trunk development in Tribolium.
MATERIALS AND METHODS
Tc-btd sequence was identified in the Tribolium genome (GenBank accession number: NM_001114320; Tc number: TC011696). A cDNA fragment of Tc-btd spanning the Btd box and zinc finger domains was amplified by using PCR (Tc-btd_F: 5’-GTGACTACTATGATGGCT-3’ and Tc-btd_R: 5’-TTGCTCCCCCAAAGTAAT-3’) then cloned into the pGEM(r)-T Easy Vector (Promega). The cDNA sequence is available in Genbank under the accession number of MK546416.
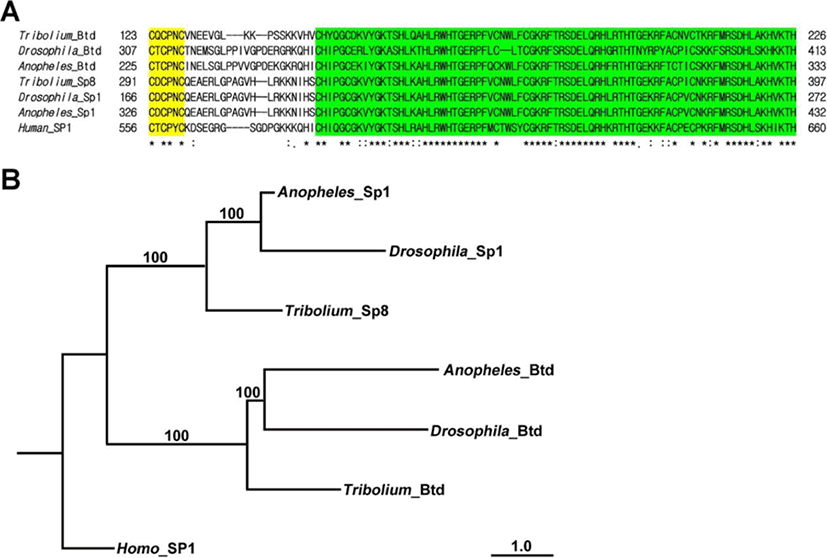
Phylogenetic analysis was carried out using the amino acid sequences of insect orthologs of Btd obtained from Genbank: (Drosophila-Btd, Drosophila melanogaster NP_511100; Anopheles-Btd, Anopheles gambiae XP_311200; Drosophila-Sp1, Drosophila melanogaster NP_727360; Anopheles-Sp1, Anopheles gambiae XP_311199; Tribolium-Sp8, Tribolium castaneum NP_001034509; Human-SP1, Homo sapiens NM_001251825). CLUSTAL Omega with default parameters was used to align the CDS of Tc-Btd with other Btd/Sp orthologs (Sievers et al., 2011). A maximum likelihood tree was constructed using the PhyML method with default parameters (Lefort et al., 2017).
Whole-mount in situ hybridization was performed as described with some modifications (Kim et al., 2013). The digoxigenin-labeled Tc-btd riboprobe was transcribed with 1μg of the pGEM(r)-T Easy Vector containing the Tc-btd cDNA fragment using T7 RNA polymerase (Ambion). Wild-type (GA2) embryos were collected in 24 hours-post-crossing, which were temporally heterogeneous. The embryos were dechorionated in 50% liquid chlorine bleach and then fixed for 15 min in 4% formaldehyde in Phosphate-Buffered Saline (PBS). The fixed embryos were dehydrated in MeOH, rehydrated in PBT (PBS containing 0.1% Tween 20), then treated with xylene to increase permeability for the riboprobes. Embryos were hybridized with Tc-btd riboprobes in hybridization buffer (50% formamide / 5X Saline Sodium Citrate Buffer (SSC) / yeast RNA at 200μg/mL / heterologous DNA at 100 μg/mL / 0.1% Tween 20). The hybridized riboprobes were detected by Anti-Digoxigenin-AP (Roche) in conjunction with BM purple (Roche). Immunochemistry was performed as described previously with a 1:5 dilution of mAb anti-En (4D9) from the Developmental Studies Hybridoma Bank (DSHB) at the University of Iowa (Patel et al., 1994). Embryos that were stained with Tc-btd riboprobe or anti-En (4D9) antibody were photographed on a BX50 compound microscope (Olympus, Melville, NY).
Parental RNAi for knocking down Tc-btd was performed as described (Bucher et al., 2002). A PCR fragment of Tc-btd that contains a T7 promoter on each end was generated with a T7-linked primer set (Tc-btd_T7F: 5’-TAATACGACTCACTATAGGGACCACCTGCACCA GGATGTAC-3’ and Tc-btd_T7R: 5’-TAATACGACTCA CTATAGGGTTCACCCCGTCACTGCTTTT-3’). dsRNA for Tc-btd was synthesized with 1μg of PCR DNA template using T7 RNA polymerase (Ambion). 500 ng/μL or 750 ng/μL of Tc-btd dsRNA were injected into wild-type (GA2) pupae. Injection buffer (0.1mM Sodium Phosphate Buffer pH 6.8, 5mM KCl) or 1 μg/μL of Tc-fushi tarazu (FTZ) dsRNA was injected as a negative control and produced no defects in the head and trunk, as reported previously (Choe et al., 2006). Embryos were collected every 48 hours for 2 weeks and then incubated at 30°C for 24 hours or for 4 days to analyze the RNAi phenotypes.
RESULTS
The Triboliumortholog of btd was identified in the Tribolium genome. This predicted 846 bp of Tc-btd that coded for a deduced protein of 282 amino acids. The deduced protein contained the C-X-C-P-X-C Btd box and C2H2 zinc finger domains (Wimmer et al., 1993). A fragment of Tc-btdcontainingthe Btd box and zinc finger domains was amplified from wild-type cDNA (Fig. 1A). Alignment of the deduced 104amino acid sequences with orthologs of Btd/Sp-family members from other insects displayedthe well-conserved Btd box and zinc finger domains, which werehighlighted in yellow and green, respectively in Fig. 1A. Zinc finger domains of Tribolium Btd showed 83.95% identity with those of Anopheles and69.62% identity with those of Drosophila. In addition, zinc finger domains of Tribolium Btd showed 91.36% identity with those of Tribolium Sp8, 92.59% identity with those of Anopheles Sp1, and 92.59% identity with those of Drosophila Sp1. Phylogenetic tree constructed using maximum likelihood analysis from the complete deduced amino acid sequences of insect Btd/Sp-family members confirmed that the fragment of Btd box and zinc finger domains we isolated from Tribolium cDNA belonged to the insect btd gene (Fig. 1B).
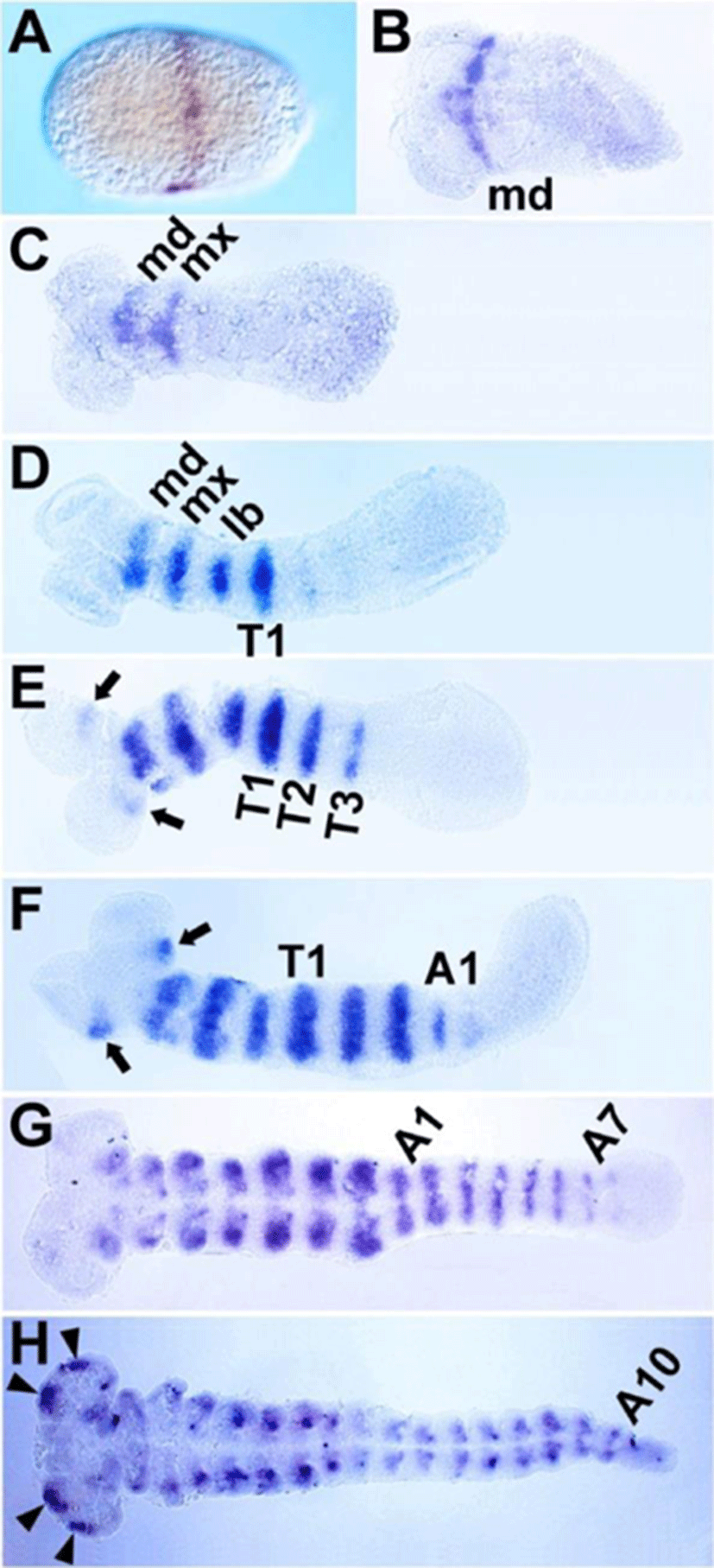
Although the expression patterns of Tc-btd during segmentation were reported previously in Tribolium(Schinko et al., 2008), we re-analyzed its expression patterns with our own riboprobe that hybridized with 629 bp of Tc-btd mRNA. The first transcripts of Tc-btd were detected as a stripe at approximately 50% egg length from the posterior pole during the blastoderm stage (Fig. 2A). When the germ rudiment formed, the first stripe was located in the mandibular (MD) segment (Fig. 2B). As the germband grew, a series of Tc-btd stripes appeared sequentially in the maxillary (MX), labial (LB), thoracic (T), and abdominal (A) segments, with their expression being maintained throughout the germband extension stages (Fig. 2C-H). When the Tc-btd stripe arose in the third thoracic (T3) segment, it appeared in the antennal (AN) segment (arrows in Fig. 2E). As the germband grew, the antennal expression became stronger than its initial expression (arrows in Fig. 2F). In the fully extended germband, new expression domains of Tc-btd were observed in the pregnathal region (arrowheads in Fig. 2H). Compared to the early expression of btd in the future head regions of Drosophila embryos (Wimmer et al., 1993), Tc-btd expression in the head appeared in later stages of head development, as reported previously (Schinko et al., 2008). The late expression of Tc-btd in the pregnathal region might imply a role for Tc-btd in maintaining a segmental identity or promoting further differentiation of the head primordium, rather than a role in patterning the head segments per se. In addition, the segmental Tc-btd stripes in the thorax and abdomen of the growing germband suggest a role for Tc-btd in the trunk segmentation.
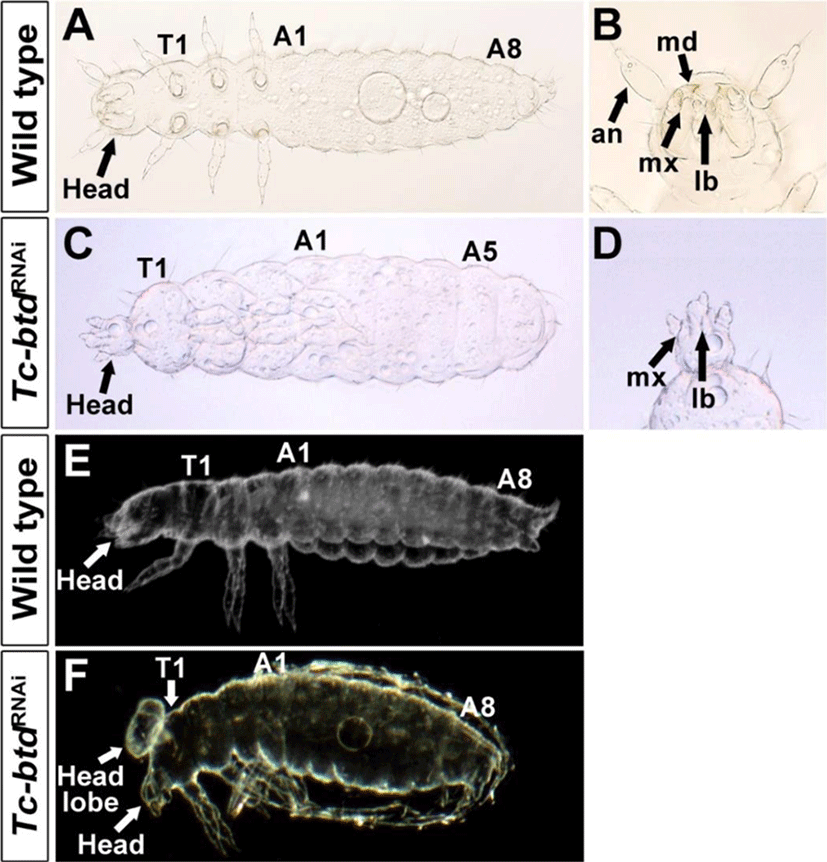
We investigated the role of Tc-btd in the head and trunk segmentation using RNAi. First, we injected 500 ng/μL of Tc-btd dsRNA into wild-type (GA2) pupae. Among 127 cuticles of Tc-btdRNAi larvae, 10.2%(13/127) showed defects in the head and trunk while the remaining 89.8%(114/127) were normal. To increase RNAi efficiency, we then injected 750 ng/μL of Tc-btd dsRNA into wild-type (GA2) pupae. In this case, 28.3%(49/173) showed similar defects in the head and trunk to those injected with 500 ng/μL of Tc-btd dsRNA. The remaining 71.7% (124/173) showed normal head and trunk segmentation. Although the penetrance of Tc-btdRNAi was quite low (10.2% to 28.3%), Tc-btdRNAi led to the reduced size of the head and the lack of several abdominal segments compared with wild types (Fig. 3A, C). In most cases, the pregnathal region including the antenna (AN), and the mandible (MD) were lost and the maxillary (MX) was abnormally shaped in Tc-btdRNAi larvae (Fig. 3B, D). However, the labium (LB) of Tc-btdRNAi larvae appeared to be normal (Fig. 3B, D). Compared with wild types, in some cases, an uncharacterized head lobe formed on the dorsal of the thorax in addition to the reduced size of the head (Fig. 3E, F). As a negative control, 1× injection buffer or 1 μg/μL of Tc-ftz dsRNA, which was previously reported to have no effects on the segmentation of larval cuticles, was injected into wild-type (GA2) pupae (Choe et al., 2006). All cuticles of 1×injection buffer injected (n=109) or Tc-ftz dsRNA injected (n=146) larvae displayed the normal head and trunk. Although it was reported previously that Tc-btd was dispensable for the development of head in Tribolium(Schinko et al., 2008), our results indicated the role of Tc-btd in the trunk segmentation as well as head development.
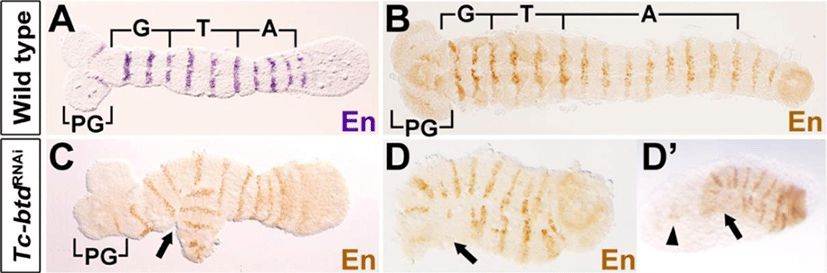
In order to verify the defects observed in the Tc-btdRNAi larval cuticles, we examined embryonic segmentation process at the stage of germband extension. To do so, we analyzed expressions of Engrailed (Tc-En) in Tc-btdRNAi embryos. In wild-type growing germbands, a series of Tc-En stripes arises sequentially in the gnathal, thoracic, and abdominal segments as well as in the antennal segment (Fig. 4A). In the growing germbands of Tc-btdRNAi embryos, we observed irregular Tc-En stripes in the gnathal region, which was bifurcated (arrow in Fig. 4C). Interestingly, the fully extended germbands of Tc-btdRNAi embryos that the segmentation was completed, were short and wide in addition to the irregular Tc-En stripes, compared with wild types (Fig. 4B, D). In particular, the head lobe including the pregnathal segments seemed to form abnormally or be absent in Tc-btdRNAi germbands (arrows in Fig. 4D). In the same Tc-btdRNAi embryo, we observed a trace of tissues (arrowhead in Fig. 4D’), which was anterior to the defective head region (arrow in Fig. 4D’) before dissecting the germband from the yolk. Such a trace of tissues was never observed in wild-type embryos. The defects in the head and trunk of Tc-btdRNAi germbands suggest that the abnormal head and abdomen of Tc-btdRNAi larval cuticles might be caused by some defects occurred in the early segmenting germbands.
DISCUSSION
We have re-analyzed expression patternsand functions of Tc-btd in Tribolium segmentation. We confirm the segmental expression of Tc-btd with single segmental periodicity during embryonic segmentation, as reported previously (Schinko et al., 2008). However, we find that Tc-btd is required for the development of the head and trunk in the later larval cuticles as well as in the early embryonic germbands, which was not revealed in the previous report (Schinko et al., 2008). This finding raises immediately two issues on the role of Tc-btd in Tribolium development.
The first issue is why the requirement of Tc-btd in the head development was not revealed in the previous study that utilized the parental and embryonic RNAi and confirmed significant knock-down of Tc-btd (Schinko et al., 2008). A reason for that could be the different genetic background of strains that has a major effect on the penetrance and severity of defects for the loss of a gene. Indeed, it has been reported that phenotypes generated with RNAi knock-down depend on the genotype of the used strain in Tribolium(Kitzmann et al., 2013). To knock downTc-btd, we utilized wild-type GA2 strain in the current study, whereas the wild-type stain used for knocking down Tc-btd was not properly documented in the previous report (Schinko et al., 2008). Thus, the different genetic background of the wild-type strains used in the previous and current studies might be a reasonable explanation for the different conclusion on the Tc-btd function in head development. However, we could not completely rule out a possibility that potential off-target effects by knock-down of Tc-btd may cause the defects in the head and trunk.
The second issue is on the effect of Tc-btd in the head and trunk development; is it necessary to pattern or shape the germbands? In order to shape the germband, first the germ rudiment forms with the condensation of blastomeres, and then extends its length fully through the combination of proliferation of cells at the posterior tip of the germ rudiment and the convergent extension of those cells (Brown et al., 1994; Cepeda et al., 2017). The head is patterned at the stage of blastoderm even before the germ rudiment forms, and the trunk is patterned at the stage of germband extension (Brown et al., 1994). The Tc-En stripes in the bifurcated head regions of the early growing Tc-btdRNAi germband suggest that somehow head patterning is achieved but the morphogenesis of the germband is disrupted. The short and wide germband with the Tc-En stripes in the fully extended Tc-btdRNAi germband shows that germband growth is disrupted, whereas somehow the pattering of the trunk segmentation occurs. In addition, compared with other Tribolium segmentation genes that are expressed in the broad domains of the early blastoderm and in the posterior regions of the growing germbands (Choe et al., 2006; Choe & Brown, 2007; Sarrazin et al., 2012; El-Sherif et al., 2012), Tc-btd seems to be expressed too late for patterning the head and trunk segmentation in the blastoderm and growing germbands, respectively. Based on the phenotypes of Tc-btdRNAi germbands and the expression patterns of Tc-btd, we propose that Tc-btd could be involved in shaping rather than in patterning the germbands at the stages of blastoderm and germband extension. In this context, the tissues of the abnormally shaped Tc-btdRNAi germbands would not be patterned and survived properly in the later stages of head and trunk development, so would lead to the loss of the pregnathal and some abdominal segments in the Tc-btdRNAi larvae. Comparative functional analysis of btd orthologs in other insects would reveal to what extent btd plays a role in patterning or morphogenesis for the development of early germbands.

