INTRODUCTION
Olive flounder (Paralichthys olivaceus) is a saltwater fish belonging to the flounder family. It is a representative species among fish cultured in Korea. Although it is a commercially important fish, productivity has declined due to diseases, increase in breeding density, deterioration of breeding environment, shortage of labor, and increase in aquaculture costs. High-density production to reduce costs has led to an increase in diseases among fish. This factor, in particular, is emerging as a major problem in olive flounder farming (Seo, 2020). In pisciculture, stocking density determines fish growth rate, feed intake rate, aquaculture productivity, and survival rate (Refstie, 1977; Rowland et al., 2006). An increase in stocking density can act as a stress factor for fish and adversely affect their growth (Holm et al., 1990; Björnsson, 1994).
Viral hemorrhagic septicemia virus (VHSV) infection has been reported to cause damage to farmed olive flounders during periods of low water temperatures in winter and spring in Korea. The infection has also been detected in wild fish (Kim et al., 2003). Infected fish show symptoms such as bleeding of internal organs, skin, and muscles; infection of the eyes, skin, gills, and fins; and abnormal symptoms such as incessant movement (Isshiki et al., 2001; Kim et al., 2009).
Immunity is an important physiological system that detects and effectively defends against external pathogens as organisms evolve and adapt to change in various environments. Immunity is divided into two categories: innate immunity (natural immunity) and adaptive immunity (acquired immunity). In fish, innate immunity is more important because the specificity between antigens and antibodies is low (Magnadóttir, 2006).
Interferons (IFNs) are a type of cytokines that are secreted either by virus-infected cells (type 1 interferons, IFN-α/β) or by engagement of T-cells and NK cells (type 2 interferons, IFN-γ) and are important against some viral infections. There is a type 3 interferon as well that is known to act as a stimulant (Lengyel, 1982; Pestka et al., 1987; Espinosa et al., 2017). The interferon regulatory factor (IRF) family plays an important role in the activation of IFN genes involved in host immune responses. At least 11 members of the IRF family (IRF1 to IRF11) have been identified in vertebrates (Yeow et al., 2000; Huang et al., 2010). Among the IRF family members, IRF3 plays an important role in inducing the expression of the type 1 IFN gene in virus-infected cells and is structurally homologous with IRF7 (Barnes et al., 2002; Huang et al., 2010). In addition, IRF3 is mainly present in the cytoplasm and is ubiquitously expressed in various tissues (Au et al., 1995). Upon viral infection, phosphorylation of specific serine residues in the C-terminal (regulatory) region induces translocation of IRF3 and the NF-κB protein complex from the cytoplasm to the nucleus. Upon translocation to the nucleus, IRF3 and NF-κB initiate transcription, resulting in the activation of the classical JAK-STAT pathway and interferon-stimulating gene factor (ISGF)-3 (Tamura et al., 2008; Wang et al., 2016). Currently, fish IRF3 has been reported in seabass (Lates calcarifer), Grass carp (Ctenopharyngodon idella), rainbow trout (Oncorhynchus Mykiss) and Red sea Bream (Pagrus major), but the results are limited (Holland et al., 2008; Xu et al., 2015; Krishnan et al., 2019; Kim et al., 2020). Therefore, in this study, the expression of IRF3 in olive flounder tissue was analyzed in different developmental stages. The expression of IRF3 in the muscle and brain tissues of fish after VHSV infection at different stocking densities was also analyzed.
MATERIALS AND METHODS
The fish used in this experiment was common olive flounder, bred at the Fish Breeding Research Center of the National Institute of Fisheries Science in a 5-ton round tank at a water temperature of 16.8°C to 20.4°C and a photoperiod of 15 h of light and 9 h of darkness. According to the growth of olive flounder, commercial feeds on various size and nutrients were feed. Rotifers were feed as the main food until 3 to 20 days after hatching (DAH), and Artemia was additionally supplied until 13 to 31 days. From the 18th DAH, the compound feed were additionally supplied, and the proportion of the compound feed was gradually increased.
From egg to 50 DAH, whole body samples of olive flounder were observed for the expression of each developmental stage. Up to 10 DAH from eggs, sample were sampled so that the amount of sample was 0.1 g, and as growth progressed, 10 sample were crushed and 0.1 g was used. The samples were placed in TRI-Solution™ (TS200-001, Bio Science Technology, Daegu, Korea) and stored at −80°C until use. To observe tissue-specific expression, muscles, brain, heart, eyes, fins, skin, gills, stomach, intestines, liver, kidneys, and spleen were extracted from 8-month-old healthy olive flounders (total length approximately 30 cm, n=3) and stored at −80°C in TRI-Solution™ until the experiment.
The purpose of this study was to analyze the expression density of IRF3 in the tissues of healthy 8-month-old olive flounder (total length approximately 30 cm, n=3) after artificial VHSV infection. The fish raised in a 5-ton tank were transferred to a 3-ton round tank the day before the experiment and were given no feed. For the experiments, the fish were divided into an experimental group and a control group. The control group was inoculated with 100 µL 1× PBS (phosphate buffered saline) and the experimental group was inoculated with the VHSV suspension (104.8 TCID50 virus/fish) (Kong et al., 2009). After inoculation, according to the standard manual for olive flounder farming (Kim et al., 2016) and the procedure outlined by Eh (2011), the breeding density was divided into high density (coverage rate 3), standard density (coverage rate 1), and low density (coverage rate 0.34). Samples were taken from the brain and muscle of olive flounders every 0, 3, 6, 9, 12 hours and 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 days. To minimize the stress of the experimental fish, they were anesthetized with 150 ppm of MS-222 (Sigma-Aldrich, St. Louis, MO, USA). Each tissue was stored at −80°C prior to the experiment (Noh et al., 2017).
Total RNA was extracted from 1 g of sample using the TRI solution protocol. Protein contamination and buffer or phenol contaminations were confirmed spectrophotometrically by measuring A260/280 and A260/230 ratios (Eon™ Microplate Spectrophotometer, BioTek, Winooski, VT, USA). Total RNA (500 ng) were synthesized using the Transcriptor First Strand cDNA Synthesis Kit (Roche, Mannheim, Germany) for oligo-d(T)18 primers. cDNA (100 ng/µL) was subjected to qRT-PCR under the following conditions: initial denaturation at 95°C for 20 s, annealing at 58°C for 30 s, and elongation at 60°C for 30 s. Internal standard control was taken as 18S rRNA and quantification was performed using the 2-ΔΔCt method (Pfaffl, 2001). IRF3 primer (forward: 5’-GAAAGCCAGGTTATCCCAGG-3’, reverse; 5’-AGATTGGCTT GGGTTCGATC-3’) and 18S rRNA primer (forward: 5’-ATGGCCGTTCTTAGTTGGTG-3’, reverse: 5’-CACACGTGATCCAGTCAGT-3’) were designed using the Primer3 program after a nucleotide sequence search through the NCBI BLAST database (Rozen & Skaletsky, 2000).
The results of this study were subjected to one-way analysis of variance followed by Duncan’s LSR (least significant range) test using the software package R version 3.0.1 (Okorie et al., 2013). All data were expressed as mean±SD (n=3). p-values <0.05 were considered to indicate statistical significance.
RESULTS
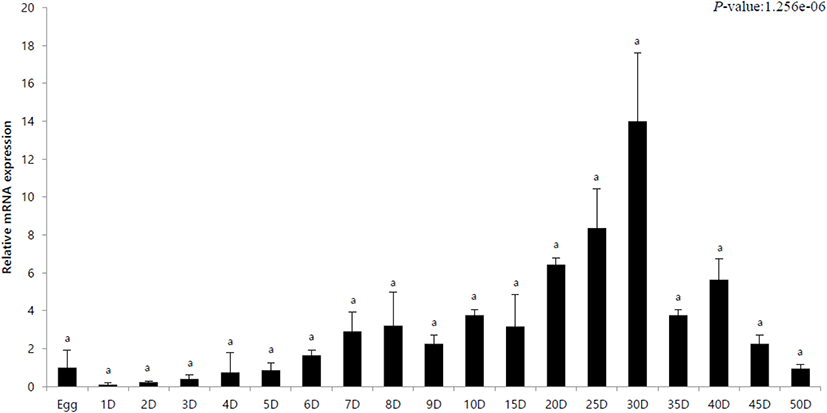
The innate immune response in olive flounder against VHSV was investigated by analyzing the expression of the IRF3 gene during the first 50 days post hatching. mRNA expression through the developmental stages was calculated using 18S rRNA primers and relative expression was calculated with respect to expression in egg (its value designated as 1). In addition, quantitative analysis was performed using IRF3-specific primers. The expression of IRF3 in hatchlings from day 1 to day 5 was lower than that in the eggs. The expression gradually increased after day 6 and was highest on day 30 (Fig. 1). After this, it gradually decreased and showed a lower expression than that in the eggs on the day 50 (Fig. 1).
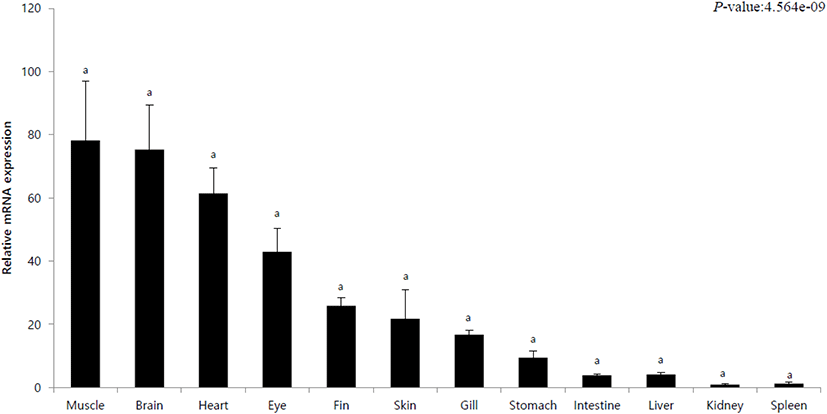
To observe the tissue-specific expression of IRF3, healthy olive flounder tissues (muscle, brain, heart, eyes, fins, skin, gills, stomach, intestine, liver, kidney, spleen) were used. Tissue-specific mRNA expression was calculated using the 18S rRNA primer, and the relative expression with respect to the expression in spleen (its value designated as 1) was investigated. In addition, quantitative analysis was performed using IRF3-specific primers. IRF3 showed the lowest expression in the spleen and the kidneys. The highest expressions were observed in the muscles and the brain (Fig. 2). In addition, high levels of expression of observed in the heart and skin of the eyes (Fig. 2). In the intestine and liver, expression was as low as 4 (Fig. 2).
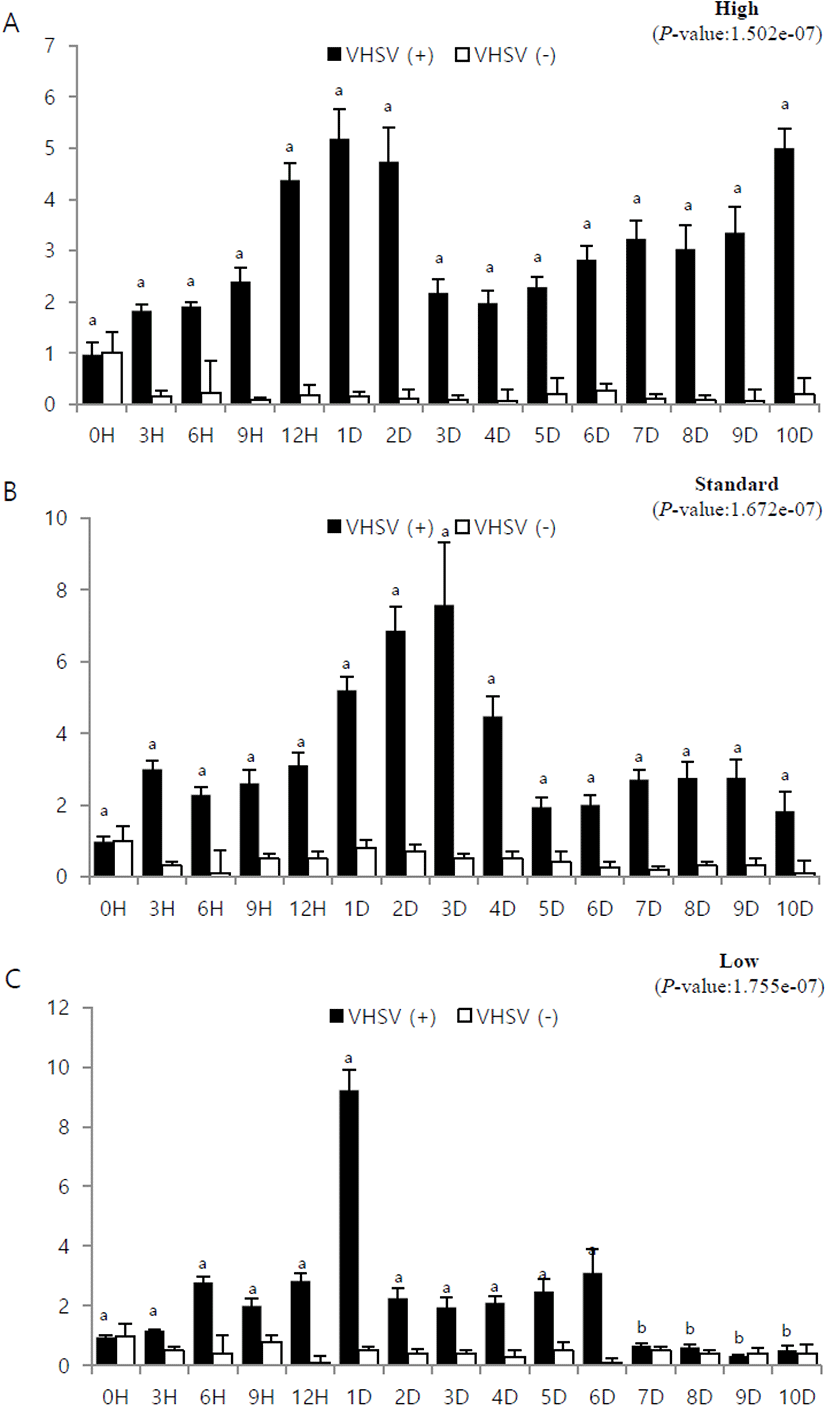
To study the immune response during viral infection with respect to the stocking density and time lapsed from infection, the mRNA expression of IRF3 was assessed using 18S rRNA primer. The relative expression was investigated with respect to expression at 0 h post-infection (its value designated as 1). In addition, quantitative analysis was performed using IRF3-specific primers. Experiments were conducted using muscle and brain tissues which, as mentioned in section 3.2, showed high IFR3 expression (Fig. 2).
In the muscle tissue from fish at high stocking density, the mRNA expression gradually increased over time and peaked at 9 h post-infection. It then decreased gradually and showed the lowest expression on day 3, after which it gradually increased again (Fig. 3A). In the muscle tissue from fish at standard stocking density, expression gradually increased until day 4, after which it decreased (Fig. 3B). In the muscle tissue from fish at low stocking density, the expression was highest at 6 h post-infection and then gradually decreased (Fig. 3C).
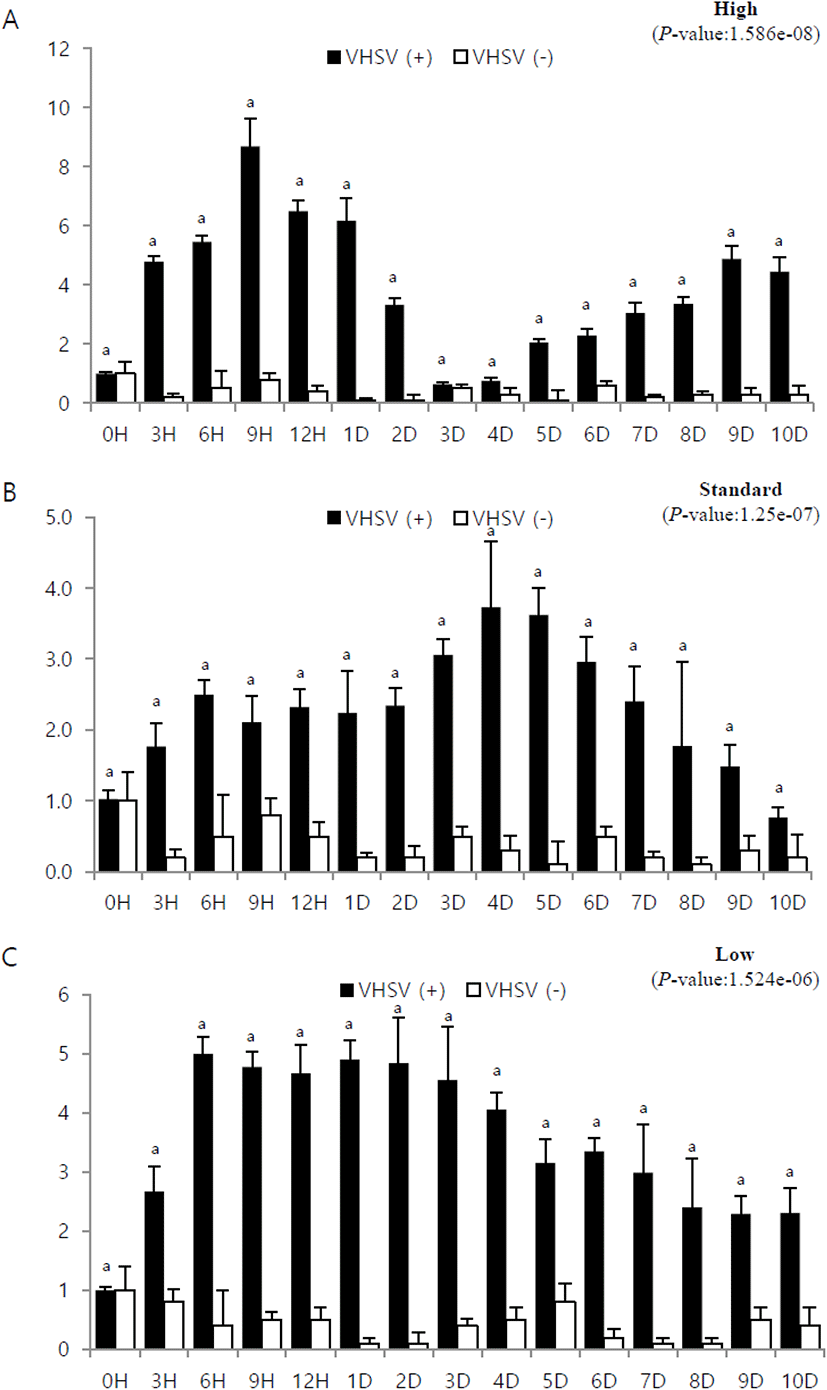
The mRNA expression in brain tissue from fish at high stocking density increased after 0 h post-infection and showed high expression on day 1. After a sharp drop on day 3, it showed a slight upward trend and then showed a high expression on day 10 (Fig. 4A). Brain tissue from fish at standard stocking density showed the highest expression on day 3 and gradually decreased thereafter (Fig. 4B). Expression in brain tissue from fish at low stocking density increased rapidly on day 1 and decreased sharply thereafter (Fig. 4C).
DISCUSSION
Although flounder is an economically important fish species, there are many problems associated with its farming, such as an increase in mortality, drug use safety, and issues with supply and demand of feed (Kim et al., 2009). VHS, one of the causes of increased mortality, is a contagious disease caused by a viral hemorrhagic sepsis virus. Factors such as fish body size, nutritional status, culture density, water temperature, and water quality affect the onset of VHS. Culture density also affects the overall breeding of farmed fish species. Fish are exposed to many pathogens from the time they are young, produce antibodies and destroy microorganisms as an innate immune response (Magnadóttir, 2006). IRF3 is a member of interferon-regulated transcription factor family and is known to play an important role in the innate immune response to viral infection (Collins et al., 2004). In this study, the innate immune response of olive flounder was quantified using IRF3 gene expression, and the tissue-specific immune response to VHSV and the immune response of VHSV on the basis of aquaculture density were investigated.
According to Han et al. (2007), on day 5, the yolk is completely absorbed, the heart and the liver develop below the esophagus, and morphogenetic differentiation of the digestive tract occurs. Between days 7 and 8 day, the stomach develops. Between days 13 and 14, the digestive tract is fully differentiated and the basic structure of the digestive tract develops (Kim, 2009). On day 30, the digestive tract is clearly divided and develops into a structure similar to that of an adult fish. The thymus, kidney, and spleen are the major lymphoid organs in teleosts. The formation of lymphocytes determines when a fish’s full immunity is achieved (Zapata et al., 2006). In this study, the expression of IRF3 gradually increased from day 6, showed the highest values on day 30 (the late larval stage), and gradually decreased by day 40 (Fig. 1). The expression of IRF3 gradually increased from the formation of immune tissues, and then decreased after the formation of immune tissues was completed. These results are similar to those of Asian seabass (Krishnan et al., 2019). It was observed that IRF3 has a higher expression in the later juvenile stage, when immune tissue is formed, than in the early juvenile stage and has a significant effect on initial immunity.
IRF3 is a member of the IRF family and is a protein-coding gene containing multiple functional regions, including nuclear export signals, DMA-binding domains, and multiple phenotype phosphorylation sites (Hiscott et al., 1999). Proteins are major transcriptional regulators of type I interferon-dependent immune responses that play an important role in innate immune responses against DNA and RNA viruses (Tanaka & Chen, 2012). The expression of IRF3 in rainbow trout (Oncorhynchus mykiss) did not show any difference in tissue-wise expression and in Asian seabass (Lates calcarife), the expression was high in the spleen and liver (Holland et al., 2008; Krishnan et al., 2019). In this experiment, the expression was approximately 80 times higher in the muscle and the brain than in the spleen (Fig. 2). Hence, the expression of IRF3 in fish tissues may vary depending on the fish species.
If the stocking density of fish exceeds a certain range, bacteria and parasites multiply rapidly, and mass death and disease may occur. In addition, high-density breeding acts as a stress factor for fish and inhibits growth, while low-density breeding reduces feed intake (Oh et al., 2013; Holm et al., 1990). Adequate aquaculture density lowers the disease incidence and high-density aquaculture increases disease incidence, so aquaculture density can be a criterion for disease incidence (Brock, 1992; Fulks & Main, 1992). Since the olive flounder is a bottom-dwelling fish, the coverage rate is used as a standard for calculating stocking density, and on a commercial scale, it is better to maintain the coverage ratio of 0.5 to 1 for better growth and survival rates (Kim et al., 2016). High density, standard density, and low density were set based on the standard manual for olive flounder farming based on the appropriate coverage rate (=1). In this experiment, IRF3 expression in olive flounder was observed on the basis of stocking density after VHSV inoculation to understand the variation in immune response. After VHSV inoculation, the expression was generally high in the muscles at low density. The expression gradually increased and then decreased in the muscles at high and standard density (Fig. 4). In addition, the expression gradually increased and then decreased in the brain at high and standard densities; however, at low density, the expression increased rapidly on day 1, after which it decreased rapidly (Fig. 3). These results conclude that the immune response of olive flounder to VHSV varies with culture density. Further investigation is needed, particularly on the immune response at low density.

