ARTICLE
Genetic Distances and Variations of Three Geographic Hairtail Populations Identified by PCR Analysis
Jong-Man Yoon†
Author Information & Copyright ▼
†Corresponding author: Jong-Man Yoon, Department of Aquatic Life Medicine, College of Ocean Science and Technology, Kunsan National University, Gunsan 573-701, Korea. Tel. : +82-63-469-1887, Fax : +82-63-463-9493, E-mail:
jmyoon@kunsan.ac.kr
Copyright © 2014 © Korean Society of Developmental Biology. All Rights Reserved.
Received: Aug 7, 2014 ; Revised: Aug 14, 2014 ; Accepted: Aug 22, 2014
ABSTRACT
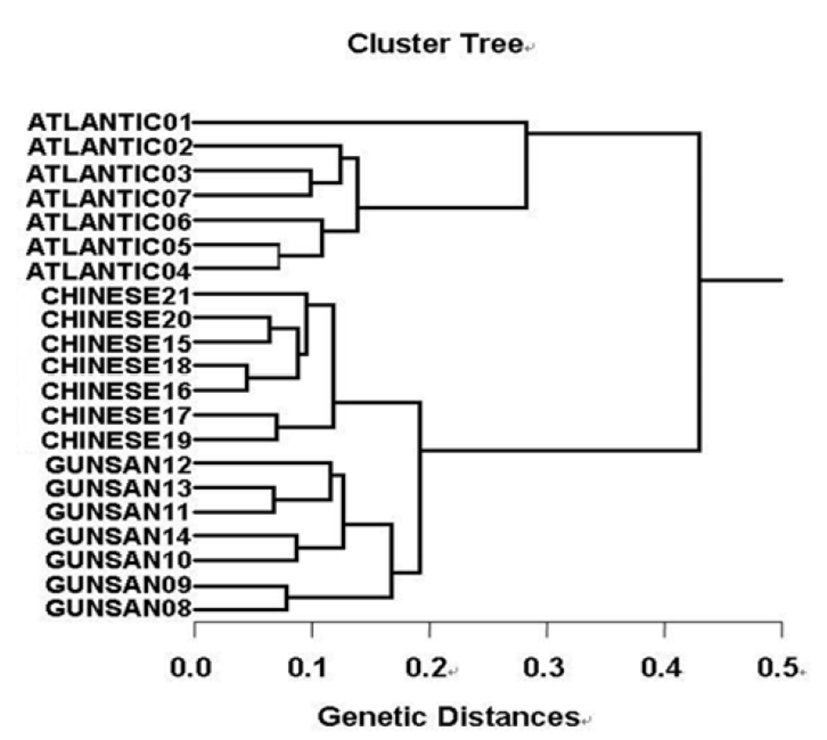
In the present study, muscle tissues were obtained separately from individuals from Atlantic hairtail population (AHP), Gunsan hairtail population (GHP) and Chinese hairtail population (CHP), respectively. The seven decamer primers were used to generate the shared loci, specific, unique shared loci to each population and shared loci by the three hairtail populations. Here, averagely, a decamer primer generated 64.7 amplified products per primer in the AHP population, 55.7 in GHP population and 56.4 in CHP population. The number of unique shared loci to each population and number of shared loci by the three populations generated by genetic analysis using 7 decamer primers in AHP, GHP and CHP population. 119 unique shared loci to each population, with an average of 17 per primer, were observed in the AHP population, and 28 loci, with an average of 4 per primer, were observed in the CHP population. The hierarchical dendrogram point out three main branches: cluster 1 (ATLANTIC 01 ~ ATLANTIC 07), cluster 2 (GUNSAN 08 ~ GUNSAN 14) and cluster 3 (CHINESE 15 ~ CHINESE 21). The shortest genetic distance displaying significant molecular difference was between individuals’ CHINESE no. 16 and CHINESE no. 18 (0.045). In the long run, individual no. 01 of the AHP population was most distantly related to CHINESE no. 19 (genetic distance = 0.430). Consequently, PCR analysis generated on the genetic data displayed that the geographic AHP population was widely separated from CHP population, while individuals of CHP population were fairly closely related to those of GHP population.
Keywords: Cluster; Euclidean genetic distance; Hairtail; Hierarchical dendrogram; Trichiurus lepturus
INTRODUCTION
Hairtails (Trichiurus lepturus) are the most popular marine products in Korea because of their taste and nutritional value, and Koreans consume them in large quantities. Hairtail, ecologically important warm water fish species, belonging to family Trichiuridae, widely distributed on the coast of the Yellow Sea, southern sea and Jeju Island in the Korean Peninsula and the several sea areas in China under the natural surroundings. However, this species of hairtail, which are acknowledged important physiologically (Koo et al., 2004), ecologically (Park et al., 2002) as well as biologically (Kim et al., 1998) are not genetically studied or researched like other finfishes. Currently, imported hairtail have been altered into endemic hairtail because of high edge. In the present study, to explicate the genetic distances and differences among geographical hairtail populations, the author accomplished a clustering analysis of three hairtail populations collected from Atlantic, Korea and Chinese site.
MATERIALS AND METHODS
Muscle tissues were obtained separately from individuals from Atlantic hairtail population (AHP), Gunsan hairtail population (GHP) and Chinese hairtail population (CHP), respectively. The muscle was collected in sterile tubes, immediately placed on dry ice, and stored at –40°C until the genomic DNA extraction. PCR analysis was performed on DNA samples extracted from a total of 21 individuals using seven decamer primers. Genomic DNA/purification was extracted and purified under the conditions described previously (Yoon & Kim, 2004). The DNA pellets were incubation-dried for 2 hrs, held at –40°C until analysis, and then dissolved in the TE buffer (10 mM Tris-HCl, pH 8.0; 1 mM EDTA). The concentrations of the extracted genomic DNA samples were estimated based on the absorbance at 260 nm by a spectrophotometer (Beckman Coulter, Buckinghamshire, UK). Seven primers (BION-02, BION-03, BION-04, BION-08, BION-09, BION-13 and BION-17) were shown to generate the shared loci, specific loci, unique shared loci to each population and shared loci by the three hairtail populations which could be obviously scored. The degree of variability was calculated by use of the Dice coefficient (F), which is given by the formula: F = 2 nab / (na+nb), where nab is the number of bands shared between the samples a and b, na is the total number of bands for sample a and nb is the total number of bands for sample b (Jeffreys & Morton, 1987; Yoke-Kqueen & Radu, 2006). Euclidean genetic distances within- and between-population were also calculated by complete linkage method with the support of the hierarchical dendrogram program Systat version 13 (SPSS Inc., Chicago, IL, USA).
RESULTS AND DISCUSSION
Genomic DNAs isolated from individuals obtained in Atlantic hairtail population, Gunsan hairtail population and Chinese hairtail population, respectively, were amplified at several times by PCR reactions. The amplified products were separated and/or purified by agarose gel electrophoresis (AGE) with oligonucleotide decamer primers and stained by ethidium bromide. The complication of the banding patterns was varied outstandingly between the primers and three regions, as shown in Fig. 1. The bandsharing value between individuals no. 04 and no. 05 was 0.871 within the AHP population, which was the highest value identified among the three populations (Table 1). The bandsharing value between individual’s no. 02 of AHP population and no. 12 of GHP population was 0.565, which was the lowest observed. In the present study, averagely, a decamer primer generated 64.7 amplified products per primer in the AHP population, 55.7 in GHP population and 56.4 in CHP population, as demonstrated in Table 2. Here, the seven decamer primers BION-02, BION-03, BION-04, BION-08, BION-09, BION-13 and BION-17 were used to generate the shared loci, specific, unique shared loci to each population and shared loci by the three populations, as illustrated in Table 3. The number of unique shared loci to each population and number of shared loci by the three populations generated by genetic analysis using 7 decamer primers in AHP, GHP and CHP population. 119 unique shared loci to each population, with an average of 17 per primer, were observed in the AHP population, and 28 loci, with an average of 4 per primer, were observed in the CHP population. As regards average bandsharing value (BS) results, individuals from AHP population (0.857) exhibited higher bandsharing values than did individuals from GHP population (0.792) (P<0.05), as demonstrated in Table 4.
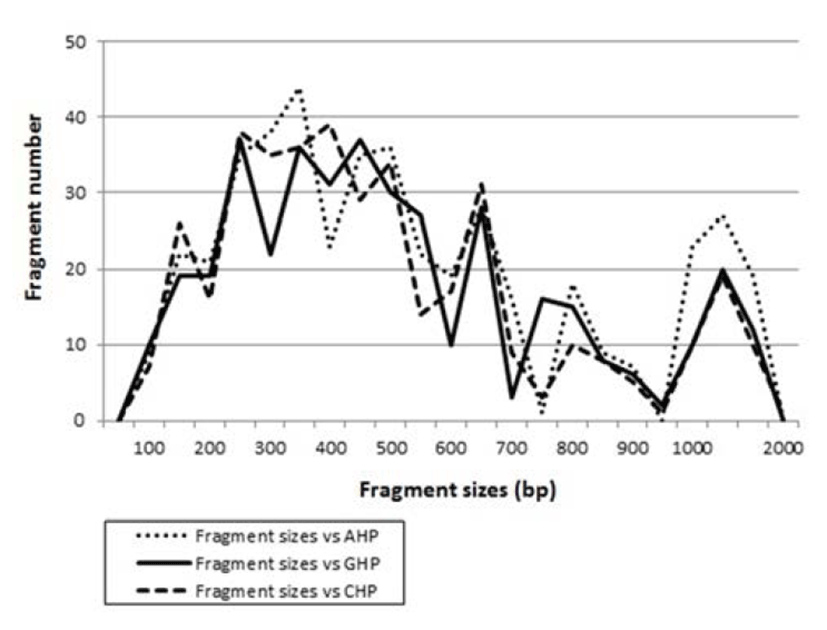
Many researchers studied the sizes of DNA fragments in the PCR profiles of five species of Eastern Pacific abalone (genus Haliotis) (Muchmore et al., 1998), black tiger shrimp (Penaeus monodon) (Tassanakajon et al., 1998), shrimp populations (Yoon & Kim, 2003) and deep sea lobster (Puerulus sewelli) (Park et al., 2005). DNA fragments attained by seven decamer primers ranged in size from 50 bp to 2,200 bp in the hairtail, as displayed in Fig. 2. The higher fragment sizes (>1,000 bp) are much more observed in the AHP population, as shown in Fig. 2. The hierarchicaldendrogram point out three main branches: cluster 1 (ATLANTIC 01 ~ ATLANTIC 07), cluster 2 (GUNSAN 08 ~ GUNSAN 14) and cluster 3 (CHINESE 15 ~ CHINESE 21), as shown in Fig. 3. The shortest genetic distance displaying significant molecular difference was between individuals’ CHINESE no. 16 and CHINESE no. 18 (0.045). In the long run, individual no. 01 of the AHP population was most distantly related to CHINESE no. 19 (genetic distance = 0.430). The genetic distance between the Indian Ocean lobster and the Korean slipper lobster species ranged between 0.040 and 0.612 (Park et al., 2005).
In other fish and invertebrates, cluster analysis of the pairwise species matrix generated from genetic data showed that geographically close populations of blacklip abalone tended to cluster together (Huang et al., 2000). A phylogenetic tree was constructed using UPGMA cluster analysis based on a total of 3,744 distinguishable fragments in gynogenetic clones from the silver crucian carp, Carassius auratus gibelio Block (Zhou et al., 2000). The interpopulation similarity indices and genetic distance values indicated that the Jamuna-Padma population pair of the Indian major carp (Catla catla) was genetically closer than the Halda-Jamuna and the Halda-Padma population pairs, which agreed with the geographical distances between them (Islam et al., 2005). They suggested that this genetic technique could be used to discriminate between different river populations of major carp. Generally speaking, using a variety of decamer primers, PCR has been applied to identify polymorphic/specific markers particular to line, species and geographical population, as well as genetic diversity/polymorphism in diverse species of organisms (McCormack et al. 2000; Yoon & Kim 2004). Consequently, PCR analysis generated on the genetic data displayed that the geographic AHP population was widely separated from CHP population. Reversely, individuals of CHP population were fairly closely related to those of GHP population, as shown in the hierarchical dendrogram of genetic distances. DNA fragments identified in this study may be useful as a DNA marker. From what has been said above, the potential of genetic analysis to identify diagnostic markers for the identification of three hairtail populations has been demonstrated.
ACKNOWLEDGEMENTS
Particular thanks go to Jun-Hyub Jeon and my laboratory colleagues for their assistance in sample collection and their assistance with the technique of PCR analysis and statistical analysis. The author thanks also to referees who assisted with detailed and insightful correction.
REFERENCES
Huang BX, Peakall R, Hanna PJ. Analysis of genetic structure of blacklip abalone (
Haliotis rubra) populations using RAPD, minisatellite and microsatellite markers. Mar Biol. 2000; 136 p. 207-216.

Islam MS, Ahmed ASI, Azam MS, Alam MS. Genetic analysis of three river populations of
Catla catla (HAMILTON) using randomly amplified polymorphic DNAs markers. Asian-Aust J Anim Sci. 2005; 18 p. 453-457.

Jeffreys AJ, Morton DB. DNA fingerprints of dogs and cats. Anim Genet. 1987; 18 p. 1-15.

Kim SH, Lee YD, Rho HK. The study on the fisheries biological feature of hairtail,
Trichiurus lepturus from the Cheju Strait. J Korean Fish Soc. 1998; 31(1) p. 17-25.

Koo JG, Jee JH, Kim JM, Kang JC. Effect of water temperature on survival, growth and intermolt period of tiger crab,
Orithyia sinica (Linnaeus) larvae. J Fish Pathol. 2004; 17 p. 139-144.

McCormack GC, Powell R, Keegan B. Comparative analysis of two populations of the brittle star
Amphiura filiformis (Echinodermata: Ophiuroidae) with different life history strategies using RAPD markers. Mar Biotechnol. 2000; 2 p. 100-106.

Muchmore ME, Moy GW, Swanson WJ, Vacquier VD. Direct sequencing of genomic DNA for characterization of a satellite DNA in five species of Eastern Pacific abalone. Mol Mar Biol Biotechnol. 1998; 7 p. 1-6.

Park CS, Lee DH, Hwang KS. Distribution and migration of hairtail,
Trichiurus lepturus in Korean waters. J Korean Soc Fish Res. 2002; 5 p. 1-11.

Park SY, Park JS, Yoon JM. Genetic differences and variations in slipper lobster (
Ibacus ciliatus) and deep sea lobster (
Puerulus sewelli) determined by RAPD analysis. Korean J Genet. 2005; 25 p. 307-317.

Tassanakajon A, Pongsomboon S, Jarayabhand P, Klinbunga S, Boonsaeng V. Genetic structure in wild populations of black tiger shrimp (
Penaeus monodon) using randomly amplified polymorphic DNA analysis. J Mar Biotech. 1998; 6 p. 249-254.

Yoke-Kqueen C, Radu S. Random amplified polymorphic DNA analysis of genetically modified organisms. J Biotechnol. 2006; 127 p. 161-166.

Yoon JM, Kim GW. Genetic differences between cultured and wild penaeid shrimp (
Penaeus chinensis) populations analysed by RAPD-PCR. Korean J Genet. 2003; 25 p. 21-32.

Yoon JM, Kim JY. Genetic differences within and between populations of Korean catfish (
S. asotus) and bullhead (
P. fulvidraco) analysed by RAPD-PCR. Asian-Aust. J Anim Sci. 2004; 17 p. 1053-1061.

Zhou L, Wang Y, Gui JF. Analysis of genetic heterogeneity among five gynogenetic clones of silver crucian carp,
Carassius auratus gibelio Block, based on detection of RAPD molecular markers. Cytogenet Cell Genet. 2000; 88 p. 133-139.

Figures and Tables
Fig. 1.
PCR-based electrophoretic profiles of Atlantic hairtail population (lane 1-7), Gunsan hairtail population (lane 8-14) and Chinese hairtail population (lane 15-21), respectively, amplified by oligonucleotides primers BION-02 (A), BION-03 (B), BION-04 (C), BION-08 (D), BION-09 (E), BION-13 (F) and BION-17 (G). Each lane shows different individual DNA samples used. Amplified products were electrophoresed on a 1.4% agarose gel and detected by staining with ethidium bromide. 100 bp ladder markers were utilized as a DNA molecular size marker (M).
Download Original Figure
Fig. 2.
Distribution of fragment sizes of Atlantic hairtail population (AHP), Gunsan hairtail population (GHP) and Chinese hairtail population (CHP), respectively. Solid lines: GHP population. Dotted lines: AHP population. Thick dotted lines: CHP population. The fragment numbers in each size interval have been computed from the pooled fragments obtained with all the primers. The higher fragment sizes (>1,000 bp) are much more ob-served in the AHP population.
Download Original Figure
Fig. 3.
Hierarchical dendrogram of genetic distances, accomplished from three hairtail populations. The relationship among different individuals from Atlantic hairtail population (ATLANTIC), Gunsan hairtail population (GUNSAN) and Chinese hairtail population (CHINESE) was created according to the bandsharing values and similarity matrix.
Download Original Figure
Table 1.
Similarity matrix including bandsharing values (BS) calculated using Nei and Li's index of the similarity of individuals from Atlantic hairtail population, Gunsan hairtail population and Chinese hairtail population, respectively
| Bandsharing values of hairtail |
| from Atlantic |
│ |
from Gunsan |
│ |
from Chinese |
| 1 |
2 |
3 |
4 |
5 |
6 |
7 |
|
8 |
9 |
10 |
11 |
12 |
13 |
14 |
|
15 |
16 |
17 |
18 |
19 |
20 |
21 |
|
| - |
0.790 |
0.815 |
0.717 |
0.726 |
0.726 |
0.864 |
|
0.783 |
0.711 |
0.619 |
0.678 |
0.615 |
0.666 |
0.652 |
|
0.631 |
0.705 |
0.570 |
0.677 |
0.591 |
0.670 |
0.700 |
1
|
|
|
- |
0.843 |
0.828 |
0.815 |
0.815 |
0.840 |
|
0.779 |
0.691 |
0.619 |
0.760 |
0.604 |
0.703 |
0.683 |
|
0.698 |
0.701 |
0.625 |
0.726 |
0.613 |
0.664 |
0.819 |
2
|
|
|
|
- |
0.855 |
0.814 |
0.777 |
0.838 |
|
0.773 |
0.743 |
0.691 |
0.754 |
0.638 |
0.716 |
0.737 |
|
0.715 |
0.754 |
0.678 |
0.720 |
0.642 |
0.702 |
0.781 |
3
|
|
|
|
|
- |
0.871 |
0.797 |
0.792 |
|
0.721 |
0.684 |
0.594 |
0.684 |
0.571 |
0.674 |
0.625 |
|
0.711 |
0.682 |
0.635 |
0.687 |
0.677 |
0.653 |
0.751 |
4
|
|
|
|
|
|
- |
0.861 |
0.799 |
|
0.704 |
0.657 |
0.655 |
0.674 |
0.565 |
0.654 |
0.612 |
|
0.680 |
0.697 |
0.632 |
0.700 |
0.582 |
0.673 |
0.710 |
5
|
|
|
|
|
|
|
- |
0.788 |
|
0.678 |
0.638 |
0.608 |
0.662 |
0.591 |
0.649 |
0.627 |
|
0.604 |
0.693 |
0.658 |
0.692 |
0.634 |
0.621 |
0.719 |
6
|
|
|
|
|
|
|
|
- |
|
0.765 |
0.724 |
0.619 |
0.685 |
0.653 |
0.680 |
0.641 |
|
0.608 |
0.722 |
0.588 |
0.726 |
0.615 |
0.673 |
0.737 |
7
|
|
|
|
|
|
|
|
|
|
- |
0.830 |
0.677 |
0.716 |
0.702 |
0.696 |
0.702 |
|
0.690 |
0.702 |
0.583 |
0.697 |
0.582 |
0.635 |
0.666 |
8
|
|
|
|
|
|
|
|
|
|
|
- |
0.775 |
0.712 |
0.772 |
0.761 |
0.749 |
|
0.635 |
0.703 |
0.609 |
0.700 |
0.677 |
0.628 |
0.636 |
9
|
|
|
|
|
|
|
|
|
|
|
|
- |
0.665 |
0.615 |
0.663 |
0.747 |
|
0.586 |
0.658 |
0.625 |
0.630 |
0.627 |
0.614 |
0.572 |
10
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
0.738 |
0.794 |
0.703 |
|
0.752 |
0.746 |
0.688 |
0.729 |
0.633 |
0.676 |
0.653 |
11
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
0.696 |
0.691 |
|
0.658 |
0.682 |
0.624 |
0.695 |
0.685 |
0.634 |
0.630 |
12
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
0.774 |
|
0.711 |
0.688 |
0.671 |
0.725 |
0.644 |
0.698 |
0.675 |
13
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
|
0.700 |
0.717 |
0.702 |
0.732 |
0.672 |
0.683 |
0.736 |
14
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
0.788 |
0.679 |
0.793 |
0.732 |
0.797 |
0.742 |
15
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
0.725 |
0.837 |
0.704 |
0.775 |
0.747 |
16
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
0.702 |
0.754 |
0.620 |
0.667 |
17
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
0.742 |
0.687 |
0.665 |
18
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
0.694 |
0.699 |
19
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
0.738 |
20
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
21
|
Download Excel Table
Table 2.
The number of average loci per lane and specific loci by PCR analysis using 7 primers in T. lepturus in Atlantic hairtail population (AHP), Gunsan hairtail population (GHP) and Chinese hairtail population (CHP)
| Item |
No. of average loci per lane |
No. of specific loci |
| Primer |
AHP |
GHP |
CHP |
AHP |
GHP |
CHP |
| BION-02 |
9.4(66) |
8.4(59) |
7.7(54) |
10 |
31 |
26 |
| BION-03 |
11.4(80) |
9.7(68) |
9.4(66) |
45 |
19 |
31 |
| BION-04 |
8.3(58) |
9.1(64) |
7.6(53) |
30 |
36 |
25 |
| BION-08 |
4.7(33) |
4.4(31) |
4.4(31) |
26 |
17 |
24 |
| BION-09 |
10(70) |
7.9(55) |
9.1(64) |
7 |
13 |
8 |
| BION-13 |
12.1(85) |
10.7(75) |
9.7(68) |
29 |
54 |
47 |
| BION-17 |
8.7(61) |
5.4(38) |
8.7(61) |
26 |
31 |
54 |
| Total No. |
64.7(453) |
55.7(390) |
56.7(397) |
173 |
201 |
215 |
| Average no. per primer |
64.7 |
55.7 |
56.4 |
24.7 |
28.7 |
30.7 |
Download Excel Table
Table 3.
The number of unique loci to each population and number of shared loci by the three populations generated by PCR analysis using 7 decamer primers in Atlantic hairtail population (AHP), Gunsan hairtail population (GHP) and Chinese hairtail population (CHP), respectively
| Item |
No. of unique loci to each population |
No. of shared loci by the three populations |
| Primer \ Population |
AHP |
GHP |
CHP |
Three populations (7 individuals per population) |
| BION - 02 |
21 |
0 |
0 |
28 |
| BION - 03 |
7 |
21 |
7 |
28 |
| BION - 04 |
7 |
7 |
7 |
21 |
| BION - 08 |
0 |
7 |
0 |
7 |
| BION - 09 |
21 |
0 |
14 |
42 |
| BION - 13 |
35 |
0 |
0 |
21 |
| BION - 17 |
28 |
0 |
0 |
7 |
| Total no. |
119 |
35 |
28 |
154 |
| Average no. per primer |
17 |
5 |
4 |
8.6 |
Download Excel Table
Table 4.
Multiple comparisons of average bandsharing values among Atlantic hairtail population (AHP), Gunsan hairtail population (GHP) and Chinese hairtail population (CHP), respectively, were generated according to the bandsharing values
| Population |
AHP |
GHP |
CHP |
| AHP |
0.857±0.042c |
0.672±0.056ab |
0.676±0.052a |
| GHP |
- |
0.792±0.053ab |
0.667±0.047ab |
| CHP |
- |
- |
0.796±0.047a |
Download Excel Table