Short communication
Genetic Distances of Paralichthys olivaceus Populations Investigated by PCR
Jong-Man Yoon
1
,
†
Author Information & Copyright ▼
1Dept. of Aquatic Life Medicine, College of Ocean Science and Technology, Kunsan National University, Gunsan 54150, Korea
†Corresponding Author : Jong-Man Yoon, Dr. Prof., Dept. of Aquatic Life Medicine, College of Ocean Science and Technology, Kunsan National University, Gunsan 54150, Korea. Tel: +82-63-469-1887, E-mail:
jmyoon@kunsan.ac.kr
© Copyright 2018 The Korean Society of Developmental Biology. This is an Open Access article distributed under the terms of the
Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits
unrestricted non-commercial use, distribution, and reproduction in any
medium, provided the original work is properly cited.
Received: Jul 27, 2018 ; Revised: Aug 22, 2018 ; Accepted: Sep 05, 2018
Published Online: Sep 30, 2018
ABSTRACT
The author carried out PCR-based genetic platform to investigate the hierarchical polar dendrogram of Euclidean genetic distances of one bastard halibut population, particularly for Paralichthys olivaceus, which was further connected with those of the other fish population, by involving with the precisely designed oligonucleotide primer sets. Eight oligonucleotides primers were used generating excessively alterating fragments, ranging in size of DNA bands from larger than approximately 100 bp to less than 2,000 bp. As regards average bandsharing value (BS) results, individuals from Hampyeong population (0.810) displayed lower bandsharing values than did individuals from Wando population (0.877). The genetic distance between individuals approved the existence of close relationship in the cluster II. Relatively, individuals of one bastard halibut population were fairly related to that of the other fish population, as shown in the polar hierarchical dendrogram of Euclidean genetic distances. The points of a noteworthy genetic distance between two P. olivaceus populations demonstrated this PCR procedure is one of the quite a few means for individuals and/or populations biological DNA investigates, for species security and proliferation of bastard halibut individuals in coastal region of the Korea.
Keywords: Euclidean genetic distances; Polar dendrogram; Bastard halibut population
INTRODUCTION
Paralichthys olivaceus is commercially important teleost species, belonging to family Paralichthyidae, order Pleuronectiformes, broadly distributed on the seashore of the Yellow Sea, southern sea and the Jeju Island of Korea, Chinese sea and Japanese sea. In the environment, the fishes inhabit the benthic flats consisting of a lot of sand and slime. Like other fishes, the rate at which the fish grows, is greatly influenced by water quality. The outer body color of this fish is yellowish and/or body color is black and grey. The color of the abdomen is yellowish brown or light white. Mainly, there are marked shifts of the fish weight, size, color and shape in P. olivaceus in keeping with the ecological surroundings of habitat such as prey, rock crystal, water temperature, feed and harsh period. The bastard halibut is environmentally and biologically very important fishes in the Korea. However, this kind of finfish, which are well-known important environmentally (Bae et al., 2017), physiologically (Kim et al., 2018), histopathologically (Kim et al., 2017), as well as aquaculturally (Lee & Yoo, 2016) are not genetically and/or molecular-biologically studied comparable other fishes. There is a necessity to understand the genetic traits and composition of this finfish population in order to evaluate exactly the patent genetic significance. PCR-based molecular research methods have been applied to study the genetic characters of various finfishes and shellfishes (Partis & Wells, 1996; Callejas & Ochando, 1998; Tassanakajon et al., 1998; Muchmore et al., 1998; McCormack et al., 2000; Zhou et al., 2000; Chenyambuga et al., 2004; Yoon & Park 2002; Islam et al., 2005; Oh & Yoon, 2014). Generally, the markers peculiar to the species, the genus or the geographical populations have been applied for the individuals and species, hybrid parentage and for the monitoring of DNA markers. Here, to clarify the Euclidean genetic distances in bastard halibut, the author undertook the clustering analyses of two geographical populations of bastard halibut (P. olivaceus) raising in the Hampyeong and Wando, respectively.
MATERIALS AND METHODS
PCR analysis was accomplished on DNA samples extracted from a total of 22 individuals using eight oligonucleotides primers. DNA extraction should be performed along with the separation and extraction methods (Yoon & Kim, 2004). 600 µL of chloroform was added to the mixture and then inverted (no phenol). After quite a few washing, the lysis buffer Ⅰ (155 mM NH4Cl; 10 mM KHCO3; 1 mM EDTA) was augmented to samples, and the mixture tubes were gently upset. The precipitates obtained were centrifuged and suspended with lysis buffer II (10 mM Tris-HCl, pH 8.0; 10 mM EDTA; 100 mM NaCl; 0.5% SDS) and added 15 µL proteinase K solution (10 mg/mL). After incubation, there was added 300 µL of 3 M NaCl and gently pipetted for a few of min. Added not phenol, 600 µL of chloroform were added to the mixture and then inverted. Ice-cold 70% EtOH was added, and then the samples were centrifuged at 19,621 g for 5 min to extract the DNA from the lysates. The concentration of the extracted genomic DNA was measured with the optical density (OD) at 260 nm by a spectrophotometer (Beckman Coulter, Buckinghamshire, UK). The DNA pellets were then incubation-dried for more than 12 hours, maintained at -70℃ until needed and then dissolved in the distilled water. The DNA amplification was performed in 25 µL containing 10 ng of template DNA, 20 µL premix (Bioneer Co., Daejeon, Korea) and the 1.0 unit primer. Amplification products were separated by 1.4% agarose (Bioneer Co., Daejeon, Korea) gel electrophoresis with TBE (90 mM Tris, pH 8.5; 90 mM borate; 2.5 mM EDTA). The 100 bp DNA ladder (Bioneer Co., Daejeon, Korea) was used as DNA molecular weight marker. The agarose gels electrophoresed were stained with ethidium bromide (Song & Yoon, 2013). The electrophoresed agarose gels were illuminated by ultraviolet rays, and photographed using a photoman direct copy system (PECA Products, Beloit, WI, USA). The oligonucleotides primer were acquired from Operon Technologies, USA. OPA-02 (5’-TGCCGAGCTG-3’), OPA-07 (5’-GAAACG GGTG-3’), OPA-18 (5’-AGGTGACCGT-3’), OPA-20 (5’-GTTGCGATCC-3’), OPB-08 (5’-GTCCACACGG-3’), OPB-09 (5’-TGGGGGACTC-3’), OPB-15 (5’-GGAGGG TGTT-3’), and OPB-17 (5’-AGGGAACGAG-3’) were displayed to yield the bandsharing values and genetic distances of the two bastard halibut populations. PCR was carried out using programmable DNA Thermal Cycler Cycler (MJ Research Inc., Waltham, MA, USA). Similarity matrix including bandsharing values between dissimilar individuals in the two P. olivaceus populations, was generated allowing formula of Jeffreys and Morton (1987) and Yoke-Kqueen and Radu (2006). A hierarchical clustering tree was accumulated using similarity matrices to yield a dendrogram, which was supported by the Systat version 10 (SPSS Inc., Chicago, IL, USA).
RESULTS AND DISCUSSION
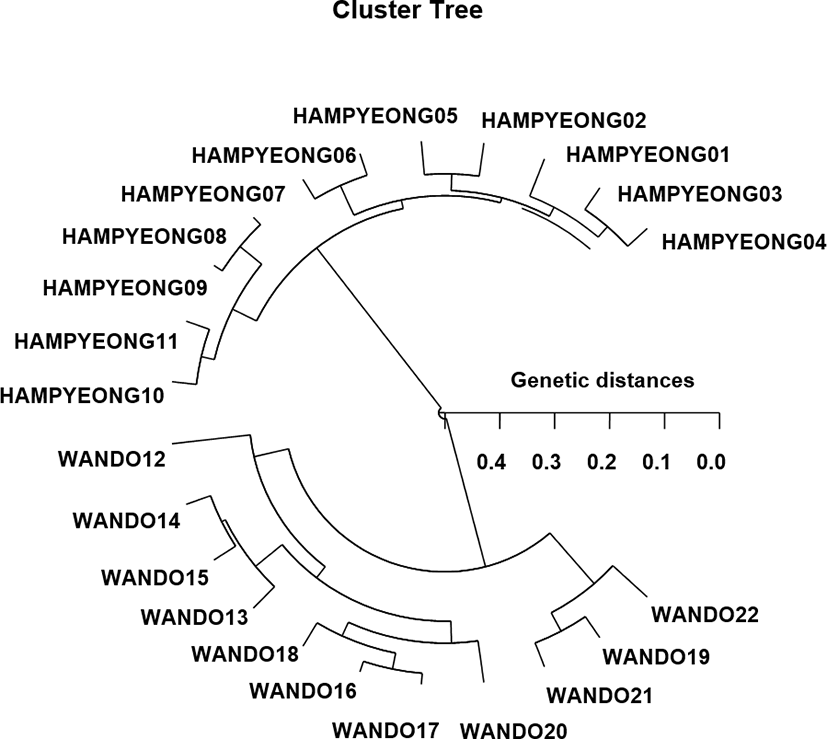
In this study, the bandsharing value, which is based on the presence or absence of amplified fragments, was utilized to calculate similarity indices in two bastard halibut populations, as demonstrated in Table 1. Here, the complexity of the banding patterns varied dramatically between the primers from the two finfish populations. The similarity matrix, which was bas ed on the average bandsharing value of all the samples, ranged from 0.677 to 0.987 in the Hampyeong population and 0.757–0.980 to the Wando population. The bandsharing value between individuals no. 08 and no. 09 within the Hampyeong population (P. olivaceus) was 0.987, which was the highest value identified among the two populations. As regards average bandsharing value (BS) results, individuals from Hampyeong population (0.810) displayed lower bandsharing values than did individuals from Wando population (0.877). The average bandsharing value reported by this study is similar to the value ported for Spanish barbell species (0.71–0.81) (Callejas & Ochando, 1998). The average bandsharing value recorded in our study is also higher than the average value between the bullhead population (0.504± 0.115) (Yoon & Kim, 2004). In the present study, the hierarchical polar dendrogram obtained by the eight oligonucleotides primers designates two genetic clusters: cluster 1 (HAMPYEONG 01, 02, 03, 04, 05, 06, 07, 08, 09, 10, and 11) and cluster 2 (WANDO 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, and 22) (Fig. 1). Among the twenty-two fishes, the shortest genetic distance that demonstrated significant molecular differences was between individuals fish no. 09 and no. 08 from the cluster I (genetic distance=0.018), while the longest genetic distance among the twenty-two individuals that exhibited significant molecular differences was between individuals fish no. 05 and clam no. 20 (genetic distance=0.489). Relatively, individuals of cluster I were greatly closely related to that of cluster II, as shown in the polar dendrogram of Euclidean genetic distances. The genetic distance between individuals approved the existence of close relationship in the cluster II. The values of the pairwise comparisons of unbiased genetic distance between the populations of the Indian major carp (Catla catla) from the combined data for the four primers, ranged from 0.025 to 0.052 (Islam et al., 2005). They reported that the Padma and the Jamuna populations were separated from each other with the lowest genetic distance (D= 0.025). From what has been said above, the prospective of this research method in determining the diagnostic markers for the breed, stock, species, genus and geographic population identification in teleost (Mamuris et al., 1999; Diaz-Jaimes & Uribe-Alcocer, 2003), in shellfish (Tassanakajon et al., 1998; McCormack et al., 2000; Oh & Yoon, 2014) and in livestock (Jeffreys & Morton, 1987; Gwakisa et al., 1994) has also been established. The points of a significant genetic distance between two P. olivaceus populations demonstrated this PCR means is one of the several devices for individuals and/or populations biological DNA investigates.
Table 1.
Trigonal similarity matrix containing bandsharing values calculated using Nei and Li’s index of the similarity of two bastard halibut (P. olivaceu s) populations from Hampyeong and Wando, respectively
|
|
Bandsharing values of Hampyeong population
|
Bandsharing values of Wando population
|
|
|
1
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
9
|
10
|
11
|
12
|
13
|
14
|
15
|
16
|
17
|
18
|
19
|
20
|
21
|
22
|
|
1
|
- |
0.831 |
0.816 |
0.831 |
0.76 |
0.738 |
0.801 |
0.713 |
0.724 |
0.794 |
0.740 |
0.659 |
0.560 |
0.550 |
0.605 |
0.603 |
0.623 |
0.564 |
0.568 |
0.538 |
0.592 |
0.549 |
|
2
|
|
- |
0.847 |
0.838 |
0.859 |
0.800 |
0.82 |
0.774 |
0.810 |
0.786 |
0.731 |
0.562 |
0.567 |
0.591 |
0.613 |
0.654 |
0.642 |
0.611 |
0.609 |
0.569 |
0.635 |
0.625 |
|
3
|
|
|
- |
0.906 |
0.785 |
0.768 |
0.802 |
0.719 |
0.751 |
0.707 |
0.677 |
0.572 |
0.572 |
0.532 |
0.512 |
0.546 |
0.609 |
0.582 |
0.548 |
0.548 |
0.570 |
0.557 |
|
4
|
|
|
|
- |
0.805 |
0.807 |
0.817 |
0.759 |
0.793 |
0.771 |
0.757 |
0.52 |
0.553 |
0.504 |
0.488 |
0.522 |
0.559 |
0.537 |
0.501 |
0.496 |
0.519 |
0.511 |
|
5
|
|
|
|
|
- |
0.918 |
0.86 |
0.891 |
0.831 |
0.789 |
0.751 |
0.551 |
0.554 |
0.544 |
0.560 |
0.596 |
0.614 |
0.565 |
0.586 |
0.523 |
0.584 |
0.589 |
|
6
|
|
|
|
|
|
- |
0.918 |
0.907 |
0.867 |
0.785 |
0.767 |
0.554 |
0.528 |
0.536 |
0.548 |
0.590 |
0.554 |
0.535 |
0.510 |
0.473 |
0.528 |
0.519 |
|
7
|
|
|
|
|
|
|
- |
0.845 |
0.84 |
0.827 |
0.761 |
0.504 |
0.506 |
0.468 |
0.483 |
0.544 |
0.537 |
0.516 |
0.515 |
0.477 |
0.536 |
0.526 |
|
8
|
|
|
|
|
|
|
|
- |
0.987 |
0.907 |
0.828 |
0.528 |
0.554 |
0.517 |
0.532 |
0.562 |
0.556 |
0.560 |
0.557 |
0.498 |
0.554 |
0.592 |
|
9
|
|
|
|
|
|
|
|
|
- |
0.894 |
0.815 |
0.566 |
0.541 |
0.53 |
0.545 |
0.576 |
0.568 |
0.573 |
0.569 |
0.511 |
0.567 |
0.557 |
|
10
|
|
|
|
|
|
|
|
|
|
- |
0.921 |
0.613 |
0.575 |
0.567 |
0.621 |
0.618 |
0.610 |
0.610 |
0.611 |
0.587 |
0.635 |
0.622 |
|
11
|
|
|
|
|
|
|
|
|
|
|
- |
0.568 |
0.545 |
0.575 |
0.624 |
0.589 |
0.580 |
0.556 |
0.556 |
0.557 |
0.578 |
0.568 |
|
12
|
|
|
|
|
|
|
|
|
|
|
|
- |
0.823 |
0.757 |
0.824 |
0.770 |
0.784 |
0.808 |
0.764 |
0.844 |
0.839 |
0.792 |
|
13
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
0.870 |
0.885 |
0.896 |
0.889 |
0.878 |
0.853 |
0.858 |
0.852 |
0.895 |
|
14
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
0.922 |
0.879 |
0.896 |
0.863 |
0.891 |
0.818 |
0.859 |
0.824 |
|
15
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
0.951 |
0.930 |
0.891 |
0.899 |
0.880 |
0.867 |
0.849 |
|
16
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
0.980 |
0.935 |
0.923 |
0.878 |
0.889 |
0.878 |
|
17
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
0.955 |
0.943 |
0.899 |
0.909 |
0.898 |
|
18
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
0.937 |
0.892 |
0.808 |
0.908 |
|
19
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
0.900 |
0.940 |
0.943 |
|
20
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
0.887 |
0.911 |
|
21
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
0.915 |
|
22
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
Download Excel Table
Fig. 1.
Hierarchical polar dendrogram of genetic distances obtained from two Paralichthys olivaceus populations. The relatedness between dissimilar individuals of two bastard halibut populations from cluster I (HAMPYEONG 01, 02, 03, 04, 05, 06, 07, 08, 09, 10, and 11) and cluster II (WANDO 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, and 22) generated according to the bandsharing values and similarity matrix.
Download Original Figure
Table 2.
Multiple calculations of average bandsharing values (mean±SE) between two bastard halibut populations were generated in keeping with the bandsharing values and similarity matrix.
| Population |
Hampyeong |
Wando |
| Hampyeong |
0.810±0.009 a |
0.560±0.004 b |
| Wando |
- |
0.877±0.007 a |
Download Excel Table
ACKNOWLEDGEMENTS
The author would like to thank the referees who aided with thorough and shrewd revision. Particular appreciations go to our laboratory colleagues and undergraduate students for their supports for their assistance in sampling, and their helps with the preparation of PCR investigation and statistical analyses during the course of this research procedures.
REFERENCES
Bae SH, Kim KW, Kim SK, Kim JH, Kim JH. 2017; Lethal toxicity and hematological changes exposed to nitrate in flatfish,
Paralichthys olivaceus in biofloc and seawater. Environ Biol Res. 35:373-379.

Callejas C, Ochando MD. 1998; Identification of Spanish barbel species using the RAPD technique. J Fish Biol. 53:208-215.

Chenyambuga SW, Hanotte O, Hirbo J, Watts PC, Kemp SJ, Kifaro GC, Gwakisa PS, Petersen PH, Rege JEO. 2004; Genetic characterization of indigenous goats of sub-Saharan Africa using microsatellite DNA markers. Asian-Australas J Anim Sci. 17:445-452.

Diaz-Jaimes P, Uribe-Alcocer M. 2003; Allozyme and RAPD variation in the eastern Pacific yellowfin tuna (
Thunnus albacares). Fish Bull. 101:769-777.

Gwakisa PS, Kemp SJ, Teale AJ. 1994; Characterization of zebu cattle breeds in Tanzania using random amplified polymorphic DNA markers. Anim Genet. 25:89-94.

Islam MS, Ahmed ASI, Azam MS, Alam MS. 2005; Genetic analysis of three river populations of
Catla catla (HAMILTON) using randomly amplified polymorphic DNAs markers. Asian-Australas J Anim Sci. 18:453-457.

Jeffreys AJ, Morton DB. 1987; DNA fingerprints of dogs and cats. Anim Genet. 18:1-15.

Kim SR, Ko SM, Choi H, Park JJ. 2017; The first report of marine leech,
Austrobdella sp. parasited on the wild flounder,
Paralithytis olivaceus and histopathological characteristics of the host. J Fish Mar Sci Edu. 29:1394-1404.

Kim JY, Sung GH, Lim JJ, Suo SA, Cho YR, Kim JH. 2018; Effects of exposure to hexavalent chromium on hematological parameters and plasma components in flatfish,
Paralichthys olivaceus. Korean J Environ Biol. 36:124-130.

Lee HY, Yoo HK. 2016; Effects of various diets on growth and body composition of juvenile olive flounder,
Paralichthys olivaceus. Korean J Ichthyol. 28:200-206.

Mamuris Z, Stamatis C, Bani M, Triantaphyllidis C. 1999; Taxonomic relationships between four species of the Mullidae family revealed by three genetic methods: Allozymes, random amplified polymorphic DNA and mitochondrial DNA. J Fish Biol. 55:572-587.

McCormack GP, Powell R, Keegan BF. 2000; Comparative analysis of two populations of the brittle star
Amphiura filiformis (Echinodermata: Ophiuroidae) with different life history strategies using RAPD markers. Mar Biotechnol. 2:100-106


Muchmore ME, Moy GW, Swanson WJ, Vacquier VD. 1998; Direct sequencing of genomic DNA for characterization of a satellite DNA in five species of Eastern Pacific abalone. Mol Mar Biol Biotechnol. 7:1-6


Tassanakajon A, Pongsomboon S, Jarayabhand P, Klinbunga S, Boonsaeng V. 1998; Genetic structure in wild populations of black tiger shrimp (
Penaeus monodon) using randomly amplified polymorphic DNA analysis. J Mar Biotechnol. 6:249-254


Yoon JM, Park HY. 2002; Genetic similarity and variation in the cultured and wild crucian carp (
Carassius carassius) estimated with random amplified polymorphic DNA. Asian-Australas J Anim Sci. 15:470-476.

Yoon JM, Kim JY. 2004; Genetic differences within and between populations of Korean catfish (
S. asotus) and bullhead (
P. fulvidraco) analysed by RAPD-PCR. Asian- Australas J Anim Sci. 17:1053-1061.

Zhou L, Wang Y, Gui JF. 2000; Analysis of genetic heterogeneity among five gynogenetic clones of silver crucian carp,
Carassius auratus gibelio Block, based on detection of RAPD molecular markers. Cytogenet Cell Genet. 88:133-139