INTRODUCTION
Arkshells are among the most popular shellfish types in Korea and are a favorite food owing to their chewy flavor and healthful worth. Scapharca broughtonii is an ecologically warm-water bivalve species, being attached to the class Bivalvia, family Arcidae, and affiliates of the genus Scapharca disseminated expansively in the southern sea, Yellow sea, and several sea areas around the Korean peninsula. The breeding seasons are from July to August. The crude cover shield of this shellfish species is blackish brown, roofed with black hair, with reddish muscle in the interior. The inner feet are long, triangular, and reddish. Like other shellfish, the growth rates of arkshells are affected pointedly by the water surroundings. Above all, there are marked differences in the figure, size, body muscle color, and shell color in S. broughtonii in accordance with the environmental conditions of the habitat, such as feed availability, water quality, and various inhabitation conditions.
Nevertheless, there have been few genetic and molecular-biological surveys of these kinds of Korean bivalves, which are appreciated as notable environmentally (Kim et al., 2007; Lee et al., 2020), reproductively (Rha et al., 2010), and nutritionally (Kim et al., 2000; Kang et al., 2020) compared to other shellfish species. Therefore, it is essential to comprehend the genetic characteristics and structure of this bivalve to assess the patent genetic effects truthfully. Differences in such traits reflect the species-distinct origin and its genetic identity (Chenyambuga et al., 2004). Numerous features of shellfish (Scapharca broughtonii) meditate their preferred seawater habitat, such as rocks, bottom, and water supremacy.
Polymerase chain reaction (PCR)-based molecular analyses have been used to explore the genetic characteristics of various fish and crustacean species (Callejas & Ochando, 1998; Liu et al., 1998; Mamuris et al., 1999; Huang et al., 2000; Yoon & Kim, 2003; Kim et al., 2004; Kim et al., 2006; Oh & Yoon, 2014; Jeon & Yoon, 2015; Yoon, 2018). Markers peculiar to a geographical population, breed, species, or genus have been applied to individuals or groups to define the genetic status, assess their hybrid parentage, or appraise the usefulness of DNA markers. To begin with, cluster scrutiny of samples of arkshell acquired in the Yeosu and Jinhae city of Korea were executed to explicate the Euclidean genetic distances (GDs) between two arkshell populations.
MATERIALS AND METHODS
The extraction of genomic DNA was put into effect under conditions as previously described ( Jeon & Yoon, 2015). Before extracting the DNA, muscle samples were acquired from 11 individual arkshells of Yeosu and Jinhae, respectively, in the Korean peninsula. The samples were centrifuged at 19,621×g for 5 min to extract the DNA from the lysate. The concentration of extracted genomic DNA was estimated based on the absorbance at 260 nm using a spectrophotometer (Beckman Coulter, Buckinghamshire, UK). The DNA balls were then incubation-dried overnight at 2°C, held at −79°C, and melted in distilled water when needed.
PCR amplification was implemented in keeping with the experiment procedures by Yoon (2018). The following oligonucleotide primers (Operon Technologies, Alameda, CA, USA) were used: OPA-20 (5’-GTTGCGATCC-3’), OPD-07 (5’-TTGGCACGGG-3’), OPD-16 (5’-AGGGCGTAAG-3’), OPD-19 (5’-CTGGGGACTT-3’), and OPD-20 (5’-ACCCGGTCAC-3’). The amplification products were separated by electrophoresis in 1.4% agarose gels with TBE and using a 100 bp DNA ladder (Bioneer Co., DaeJeon, Korea) as the DNA molecular weight marker. After electrophoresis, gels were stained with ethidium bromide, illuminated with ultraviolet ray (Oh & Yoon, 2014).
The bandsharing (BS) value was gaged by the presence/absence of amplified products at the specific positions in the same gel. These oligonucleotide primers allowed the production of BS values and a calculation of the GDs of the two arkshell bivalve populations sampled. The average of within-population similarity matrix was estimated by pairwise comparison between individuals within a population (Yoke-Kqueen & Radu, 2006). Euclidean GDs were appraised within and between populations using the hierarchical clustering program Systat version 10 (SPSS, Chicago, IL, USA).
RESULTS AND DISCUSSION
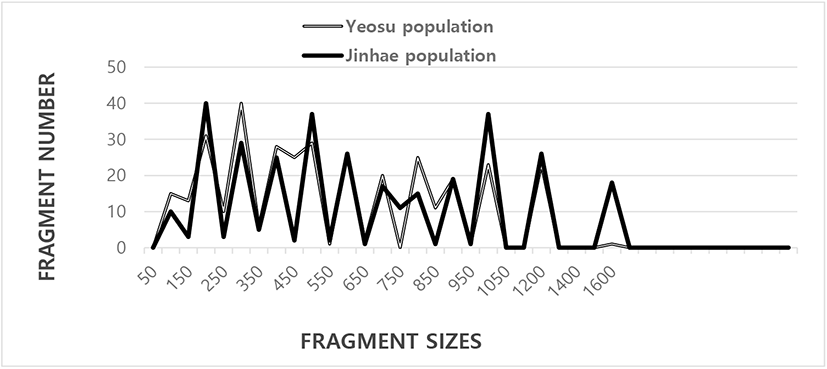
Three hundred and fifty-four and 390 DNA scorable fragments, 100 bp to 1,600 bp in size, were generated from five oligonucleotide primers for the Yeosu and Jinhae arkshell populations, respectively (Fig. 1). Zhou et al. (2000) constructed a phylogenetic tree using the unweighted pair group method (UPGMA), and 3,744 fragments in the gynogenetic clones from the silver crucian carp, Carassius auratus gibelio Block, were assessed using the arithmetic mean (UPGMA) cluster analysis method. The fragments in the bastard halibut (Paralichthys olivaceus) were acquired using four primers ranging in size from approximately 100 bp to less than 2,000 bp (Yoon, 2018). The sizes of DNA fragments obtained using random amplifications of a polymorphic DNA (RAPD) PCR-based method have been used to genetically profile the following species: blue catfish (Ictalurus furcatus) (Liu et al., 1998), blacklip abalone (Haliotis rubra) (Huang et al., 2000), oyster (Crassostrea spp.) (Kim et al., 2004), bullhead (Pseudobagrus fulvidraco) (Yoon & Kim, 2004), and populations of the eel-loach (Pangio spp.) (Siti Azizah et al., 2005). Specific primers are useful for identifying individuals based on an examination of different DNA polymorphisms (Bartish et al., 2000; Yoon & Kim, 2004; Kim et al., 2006; Oh & Yoon, 2014).
This study determined the BS values based on the existence or nonexistence of distinct fragments that used to evaluate the similarity between the samples of two arkshell populations (Table 1). The banding patterns diverged abnormally among the oligonucleotide primers from the two-arkshell population samples. The average BS values of the Yeosu and Jinhae population samples was used to determine the similarity matrix, which ranged from 0.590 to 0.862 and 0.547 to 0.893, respectively. The BS value between the individual Jinhae population sample numbers 18 and 19 was 0.893, the highest value between the two-arkshell populations sampled.
The 33 unique loci shared by each population in the Yeosu arkshell sample were generated using the OPD-20 oligonucleotides primer, approximately 200 bp, 500 bp, and 700 bp in size (Table 2). The oligonucleotide primer OPD-20 produced 55 unique loci (approximately 200 bp, 500 bp, and 700 bp in size) that were shared with each species in the Jinhae arkshell sample. Surprisingly, the primer OPD-20 generated 88 unique loci (approximately 200 bp, 300 bp, 500 bp, 600 bp, 750 bp, 800 bp, 1,200 bp, and 1,600 bp in size) that were shared with each species of Jinhae arkshell that could be used to classify each population. On the other hand, the OPD-07 primer also detected 22 loci shared by the two-arkshell populations. Those major and minor fragments were approximately 800 bp in size and comparable in all populations sampled from both populations (Table 2). The primer BION-80 detected 7 shared loci by the three venerid clam species, major and/or minor fragments of sizes 500 bp, which were matching in all samples ( Jeon & Yoon, 2015).
In the present study, the average BS value for individuals in the Jinhae arkshell samples was higher (0.700) than those in the Yeosu arkshell samples (0.692) (Table 3). The average BS values were similar to those of the Cyclina sinensis species (0.754) ( Jeon & Yoon, 2015). The BS values of the two-arkshell bivalve populations were also similar to those reported previously. The average bandsharing values in the bastard halibut (Paralichthys olivaceus) populations were 0.810±0.009 in individuals from Hampyeong population and 0.877±0.007 in individuals from Wando population, respectively (Yoon, 2018). These results were similar to those reported elsewhere: 0.802±0.010 and 0.711±0.008 for the Jeonju crayfish (JJC) and the Jeongup crayfish (JUC) populations, respectively (Kim et al., 2006). Nevertheless, the average BS value recorded in this study was higher than that reported for two oyster populations (0.282±0.008) (Kim et al., 2004).
| Population | YEOSU | JINHAE |
|---|---|---|
| YEOSU | 0.692±0.008a | 0.515±0.007b |
| JINHAE | - | 0.700±0.009a |
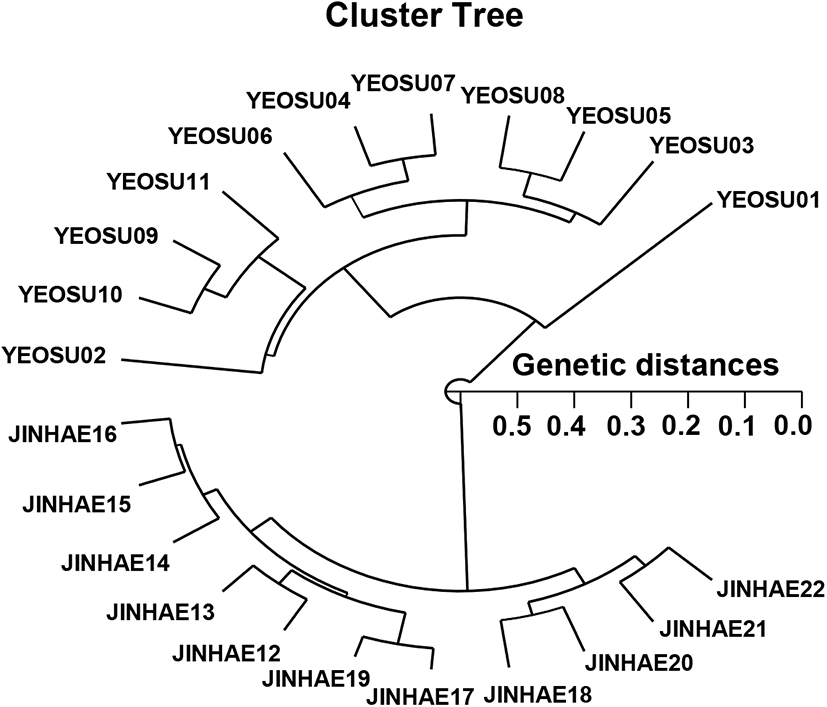
The hierarchical polar dendrogram taken using five oligonucleotide primers yielded two genetic clusters: group I (YEOSU 01, 02, 03, 04, 05, 06, 07, 08, 09, 10, and 11) and group II ( JINHAE 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, and 22) (Fig. 2). Within the twenty-two shellfish sampled, which exhibited considerable molecular differences, the individual fish no. 19 and 17 from group I (GD=0.045) showed the shortest GD. In contrast, shellfish no. 01 and 20 (GD=0.574) showed the longest GD among the twenty-two individuals that demonstrated meaningful molecular differences. The polar dendrogram of the Euclidean GDs showed that the individuals in groups I and II were distantly related (Fig. 2). Hence, there may be individual affiliations in group I. The Euclidean GDs between two populations of bastard halibut (Paralichthys olivaceus) were determined to be 0.018–0.489 from the pooled data for the eight primers used (Yoon, 2018). This technique might help define the diagnostic markers related to the breed, species, genus, and geographic population identification in finfish and crustaceans (Smith et al., 1997; Klinbunga et al., 2000; McCormack et al., 2000; Yoon & Park, 2002; Yoon & Kim, 2004; Park et al., 2005; Siti Azizah et al., 2005), plant (Bartish et al., 2000; Nozaki et al., 2000) and cattle (Suprabha et al., 2005). PCR-based analysis can determine the genetic characteristics of individuals and populations, as highlighted by the substantial GD observed between the two Scapharca populations. The survey data of the genetic variations between arkshell populations provides important statistical information for the fish husbandry. Nevertheless, specific genetic markers will be needed to characterize different populations or individuals of the genus Scapharca according to geography, relate these characteristics with the morphological features, and clarify any vagueness among genera and geographical populations. Additional sampling locations will be required to describe more accurately the area where phylogeographic disorders come about in Scapharca.