INTRODUCTION
Salmonids are among the most popular fish types in Korea and are a favorite food due to their taste, flavor, and nutritional value. Salmonids belong to the family Salmonidae in the order Clupeoidea and members of the genus Oncorhynchus include commercially important fish species. Salmonids are widely distributed in Alaska, Canada, Australia, New Zealand, South America, and the Far East. During the winter, salmonids are widely distributed in the cold-water streams and rivers draining steep mountains and flowing through the deep valleys of Gangwon-do and Chungcheongbuk-do provinces of the Korean peninsula.
Like other fish, growth rates of salmonids are significantly influenced by water quality. In particular, there are marked alterations of shape, color, size, and weight in salmonids according to changes in their ecological environment, such as food availability, prey type, rock mineral, water temperature, and life stage.
The classification of finfish species is based on morphological variations in various factors, such as body size, body type, body color, head shape and structure, and fin type and placement. It is assumed that differences in such traits reflect a species-distinct origin and its genetic identity (Chenyambuga et al., 2004). Various features of rainbow trout (RT; Oncorhynchus mykiss) and masu salmon (MS; Oncorhynchus masou) reflect their preferred freshwater habitat, such as creeks, lakes or rivers, as well as the depth of water and type of prey.
Members of the genus Oncorhynchus are environmentally and biologically important fish in Korea. Moreover, Oncorhynchus species are renowned as being aquaculturally (Baik et al., 2007), histologically (Yoon, 2002), pathologically (Sohn et al., 1993), food nutritionally (Kim et al., 2014), and morphologically (Myoung & Kim, 1993; Myoung & Kim, 1996) significant. However, compared to other species, there are relatively few genetic and molecular biological studies of Oncorhynchus species in Korea. It is crucial to determine the genetic traits and relationships of Oncorhynchus species in order to assess their genetic importance correctly.
Polymerase chain reaction (PCR)-based molecular analysis approaches have been applied to study the genetic characters of various fish and crustacean species (Partis & Wells, 1996; Tassanakajon et al., 1998; Muchmore et al., 1998; Cagigas et al., 1999; McCormack et al., 2000; Zhou et al., 2000; Yoon & Park 2002; Islam et al., 2005; Song & Yoon, 2013). In general, markers peculiar to a geographical population, breed, species, or genus have been applied to individuals or groups to determine genetic status, assess their hybrid parentage, or assess the usefulness of DNA markers. In this study, to clarify the Euclidean genetic distances between two salmonid species, the author carried out cluster analyses of samples of RT and MS being cultured in the Pyeongchang county of Gangwon-do province of Korea.
MATERIALS AND METHODS
DNA extraction was performed using previously described separation and extraction methods (Oh & Yoon, 2014). Before the extraction of DNA, muscle samples were obtained from 11 individual RT and 11 from individual MS, respectively. The collected muscle was placed in sterile tubes, immediately placed in liquid nitrogen, and stored at −79°C until undergoing genomic DNA extraction. After some washing, the lysis buffer I (155 mM NH4Cl; 10 mM KHCO3; 1 mM EDTA) was added to the samples and the mixture tubes were lightly overturned. The precipitates acquired were centrifuged and suspended with lysis buffer II (10 mM Tris-HCl, pH 8.0; 10 mM EDTA; 100 mM NaCl; 0.5% SDS) and 15 μL of proteinase K solution (10 mg/mL) was added. After incubation, 300 μL of 3 M NaCl was added, and the mixture was softly pipetted for 5 min. Chloroform (600 μL) was added to the mixture and then upset. Chilled 70% EtOH was added, and the samples were centrifuged at 19,621×g for 7 min to extract the DNA from the lysate. The concentration of the extracted genomic DNA was calculated based on the absorbance at 260 nm by using a spectrophotometer (Beckman Coulter, Buckinghamshire, UK). The DNA pellets were then incubation-dried at 2°C for overnight, held at −79°C, and, when needed, melted in the distilled water. The DNA augmentation was accomplished in a 25 μL mixture comprised of 10 ng of template DNA, 20 μL premix (Bioneer, Daejeon, Korea), and 1.0 unit of primer. Amplification products were separated by performing electrophoresis in 1.4% agarose gels with TBE (90 mM Tris, pH 8.5; 90 mM borate; 2.5 mM EDTA) and using a 100 bp DNA ladder (Bioneer) as the DNA molecular weight marker. Bands were visualized by staining with EtBr (Oh & Yoon, 2014). The electrophoresed agarose gels were brightened by ultraviolet irradiation, and pictures of the gels were obtained using a Photoman Direct Copy Camera system (PECA Products, Beloit, WI, USA).
The oligonucleotide primers OPA-07 (5’-GAAACGGGTG-3’), OPA-10 (5’-GTGATCGCA G-3’), OPA-18 (5’-AGGTGACCGT-3’), OPB-07 (5’-GGTGACGCAG-3’), and OPB-20 (5’-GGACCCTTAC-3’) were obtained from Operon Technologies, USA. The use of these five primers allowed the production of bandsharing (BS) values and the calculation of genetic distances of the two salmonid species sampled. PCR was executed repeatedly by using a programmable DNA Thermal Cycler (MJ Research, Waltham, MA, USA). A similarity matrix containing the BS values of the individuals in the two sample groups was created by applying the methods described by Jeffreys and Morton (1987) and Yoke-Kqueen and Radu (2006). The intra- and inter-species Euclidean genetic distances were calculated by using the hierarchical clustering program Systat version 10 (SPSS Inc., Chicago, IL, USA). Program Systat version 10 was also utilized to acquire additional statistical analysis results such as t-test comparisons of means and standard errors.
RESULTS AND DISCUSSION
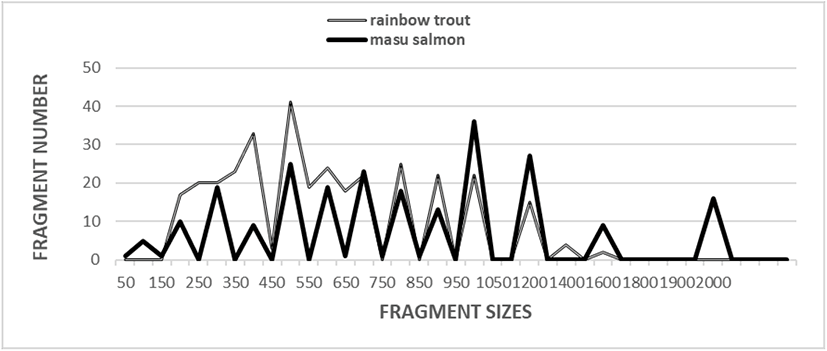
Five oligonucleotides primers were used to generate 330 scorable fragments for the RT and 234 scorable fragments for the MS population. The DNA fragments ranged in size from approximately 50 bp to more than 2,000 bp (Fig. 1). In a previous study, a phylogenetic tree was constructed using the unweighted pair group method with arithmetic mean (UPGMA) cluster analysis method to assess 3,744 distinguishable fragments in gynogenetic clones from the silver crucian carp, Carassius auratus gibelio Block (Zhou et al., 2000). In another study, the sizes of the distinguishable fragments ranged from 220 bp to 1,700 bp in four species of the Mullidae family (Mamuris et al., 1999). In black tiger shrimp, 80 bands ranging in size from 200 bp to 2,200 bp were reported (Tassanakajon et al., 1998). In the brittle star (Amphiura filiformis), the DNA fragments acquired by applying four primers ranged in size from 100 bp to 2,300 bp (McCormack et al., 2000). The use of seven oligonucleotide primers generated 317 bands in a cultured shrimp population and 385 in a wild shrimp population, and the bands ranged in size from 100 bp to 1,800 bp (Yoon & Kim, 2003a). It has been reported that a single primer can generate 9 to 15 distinct bands (Tassanakajon et al., 1998). Other researchers have also examined the sizes of DNA fragments obtained using the random amplification of polymorphic DNA (RAPD) PCR-based method to genetically profile barramundi (Lates calcarifer) (Partis & Wells, 1996), five species of Eastern Pacific abalone (genus Haliotis) (Muchmore et al., 1998), four natural Spanish populations of brown trout (Salmo trutta) (Cagigas et al., 1999), wild and cultured populations of crucian carp (Yoon & Park, 2002), marsh clams (Corbicula spp.) (Yoon & Kim, 2003b), and bastard halibut (Paralichthys olivaceus) (Yoon, 2018). Overall, specific primers have been reported to be useful for identifying individuals, with such identification based on the results obtained from examining different DNA polymorphisms (Liu et al., 1998; Park et al., 2005; Song & Yoon, 2013).
In the present study, BS values, which are determined based on the existence or nonexistence of distinct fragments, were used to evaluate the similarity between samples of two salmonid species (Table 1). At this point, the complication of the banding patterns diverse unnaturally among the oligonucleotide primers from the two salmonid samples. The similarity matrix, which was based on the average BS value of all the samples for each of the two species, ranged from 0.756 to 0.956 among the RT samples and from 0.256 to 0.927 among the MS samples. The BS value between individual RT samples number 01 and number 02 was 0.956, the highest value identified among the two species sampled.
The 66 unique shared loci to each species generated by the OPA-07 oligonucleotides primer were approximately 200 bp, 350 bp, 400 bp, 500 bp, 700 bp, and 1,200 bp in size in the RT sample (Table 2). Interestingly, the oligonucleotide primer OPB-07 produced 55 unique loci shared to each species that also pinpointed each species were approximately 250 bp, 300 bp, 400 bp, 500 bp, and 700 bp in size in the RT sample. The primer OPB-20 generated 11 unique loci shared to each species that could be used to classify each species that were approximately 1,000 bp in size in the RT sample but interestingly, primer OPB-20 also detected 22 loci shared by the two salmonid species. Those major and minor fragments were approximately 1,000 bp in size and were comparable in all sampled individuals of both species (Table 2).
Genetic variation within and among four natural Spanish populations of brown trout (Salmo trutta) was reported to be significantly higher based on results from microsatellite- and RAPD-based methods than from methods based on the analysis of enzyme loci (Cagigas et al., 1999). In the present study, the average BS value for individuals among the RT sample was higher (0.852) than that for individuals among the MS sample (0.704) (Table 3). The average BS values obtained in this study are similar to the values reported for Spanish barbel (0.71–0.81) (Callejas & Ochando, 1998). The BS values between the two salmonid species in the present study also differ from those previously reported that the average bandsharing value was 0.710±0.009 within the Korean lobster species, and 0.742±0.009 within the Indian Ocean lobster species (Park et al., 2005). However, the average BS value recorded in this study is higher than the average value reported for two oyster populations (0.282±0.008) (Kim et al. 2004). The average BS value in this study is also higher than the average value reported for bullhead populations (0.504±0.115) (Yoon & Kim, 2004).
| Species | RT | MS |
|---|---|---|
| RT | 0.852±0.006a | 0.292±0.006c |
| MS | - | 0.704±0.022b |
Each value is a result of three different experiments.
RT, rainbow trout; MS, masu salmon.
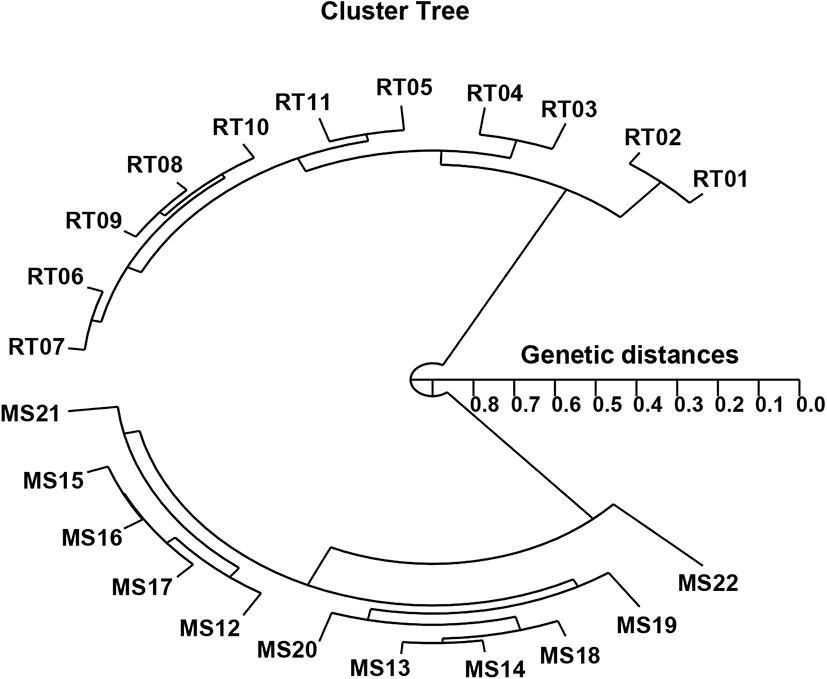
In the present study, the hierarchical polar dendrogram obtained by the use of five oligonucleotide primers produced two genetic clusters: cluster 1 (RT 01, 02, 03, 04, 05, 06, 07, 08, 09, 10, and 11) and cluster 2 (MS 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, and 22) (Fig. 2). Within the twenty-two fish sampled, that displayed significant molecular differences, the shortest genetic distance was between individual fish no. 08 and no. 09 from cluster I (genetic distance=0.034), while the longest genetic distance among the twenty-two individuals that displayed significant molecular differences was between individual fish no. 02 and fish no. 22 (genetic distance=0.846). Comparatively, individuals within cluster I were distantly related to those in cluster II, as shown in the polar dendrogram of Euclidean genetic distances (Fig. 2). The observed genetic distances between individuals reflect the presence of individual affiliations within cluster I. The pairwise comparison values of the unbiased genetic distances between populations of Indian major carp (Catla catla) from the combined data for the four primers used ranged from 0.025 to 0.052 (Islam et al., 2005). Moreover, those authors reported that the Padma and Jamuna populations were separated by the lowest observed genetic distance (D=0.025). As mentioned, the potential of the technique used in this study to define diagnostic markers related to breed, line, species, genus, and geographic population identification in finfish (Callejas & Ochando, 1998; Liu et al., 1998; Cagigas et al., 1999; Mamuris et al., 1999; Zhou et al., 2000; Yoon, 2018), crustaceans (Huang et al., 2000; McCormack et al., 2000; Kim et al., 2004; Song & Yoon, 2013), and cattle (Gwakisa et al., 1994; Chenyambuga et al., 2004; Islam et al., 2005) has been established. The observation of a noteworthy genetic distance between the samples of the two Oncorhynchus species in this study has verified that a PCR-based method is an acceptable approach to DNA-based studies into the genetic identity of individuals and populations. However, further analysis using larger sample sizes and more populations and species is needed to determine the loci peculiarities of various taxa and, specifically, to assess intra- and inter-species gene flow in Oncorhynchus.