INTRODUCTION
Till now, polymorphic bands generated by this PCR tehnique using arbitrary primers had good merits for detecting DNA similarity and diversity between life organisms (Jaiswal et al., 1998; Adams, 2000; Bartish et al., 2000; Nebauer et al., 2000; Kim et al., 2004; Yoon & Kim, 2004; Song & Yoon, 2013; Yoon, 2015). Generally, this PCR technique is one of fast and simple research methods to identify genetic difference and the polymorphism in various organisms that does not require the prior knowledge of the genomic DNA (Iyengar et al., 2000; Nicolosi et al., 2000).
Gracilaria vermiculophylla (GRV) is an economically/ cologically important seaweed species belonging to the family Gracilariaceae. GRV is only inhabited along the southern sea of Korea. This Gracilaria is widely distributed in the western sea, southern sea and Jeju Island in the Korean Peninsula as well as in Japan and Chile. Nowadays, Gracilaria is one of the most healthful algal species among the algae during the four seasons. Especially, the consumption of this alga has increased considerably in home and restaurants specializing in serving food in various ways, such as boiled rice with assorted mixtures such as red pepper spices, vegetables, fried egg, beef and/or sliced raw fish etc. The environmental necessities and the forbearances of the Gracilaria grown in different geographic locates are not known, as is Gracilaria species identification. The shape, size, color and flavour of the Graciaria blade vary according to their habitat such as the depth of the water, the strength of wave, the salinity, the water temperature, the photoperiod and the nourishment etc. Above all, the clustering analysis of the genetic distance between genera/species/populations of various fishes and invertebrates from the different geographic sites has been performed using PCR method is of small number (Tassanakajon et al., 1998; Klinbunga et al., 2000b; Yoon & Park, 2002; Kim et al., 2004; Yoon & Kim, 2004; Oh & Yoon, 2014).
As the Gracilaria farming industry is increasing considerably, the understanding of the genetics of this alga species to evaluate exactly the patent genetic characters induced by Gracilaria production operations. However, little information is known about the genetics of Gracilaria in Korea. There is a necessity to understand the genetic traits and composition of this algal species to evaluate exactly the patent genetic significance. The genetic variation, the species-specific markers and the region-specific markers in seaweeds/marine algae/marine plants have been assessed by molecular biological methods (Hong et al., 1997; Kim et al., 1997a; Kim et al., 1997b). Here, to elucidate the geetic distances and the differences in two Gracilaria species, we performed the clustering analysis and the genetic distances.
MATERIALS AND METHODS
Gracilaria vermiculophylla (GRV) and G. chorda (GRC) were obtained from Wando near the southern sea in Korea. The blades of each alga were collected, placed in the sterile tubes on ice immediately, and stored at −40° until needed. PCR analysis was performed on the blades extract of 22 individuals using different seven oligonucleotides primers. The extraction of genomic DNA was performed under conditions as described (Yoon & Kim, 2003b). After several washings, the lysis buffer Ⅰ (155 mM NH4Cl; 10 mM KHCO3; 1 mM EDTA) was added to samples, and the mixture tubes were gently inverted. The precipitates obtained were centrifuged and re-suspended with lysis buffer II (10 mM Tris-HCl, pH 8.0; 10 mM EDTA; 100 mM NaCl; 0.5% SDS) and added 15 µL proteinase K solution (10 mg/mL). After incubation, there was added 300 µL of 3 M NaCl and gently pipetted for a few of min. Without phenol, 600 µL of chloroform were added to the mixture and then inverted. DNA from the lysates was extracted by adding ice-cold 70% ethanol and centrifuged at 19,621 g for 5 min. The DNA pellets were incubation-dried for more than 2 hrs, held at −40° until analysis and then dissolved in the pure water. The concentration of the extracted genomic DNA was measured with the absorbance ratio at 260 nm by a spectrophotometer (Shimadzu, Australia; Beckman DU 600 series, UK).
The oligonucleotides primer selected were purchased from Operon Technologies, USA. The G + C content of the primers is between 60%–70%. Seven oligonucleotides primers, OPC-01 (5’-TTCGAGCCAG-3’), OPA-04 (5’-AAT CGGGCTG-3’), OPA-05 (5’-AGGGGTCTTG-3’), OPD-07 (5’-TTGGCACGGG-3’), OPD-08 (5’-GTGTGCCCCA- 3’), OPB-10 (5’-CTGCTGGGAC-3’) and OPD-16 (5’-AG GGCGTAAG-3’) were shown to generate the unique shared loci to each species and shared loci by the two species that can be scored clearly and reproducibly. Thus, we used the oligonucleotides primers to identify the genetic polymorphism, diversity and similarity of two Gracilaria species. PCR technique was performed using two programmable DNA thermal cyclers (Perkin Elmer Cetus, USA; MJ Research, Inc., USA). The DNA amplification was performed in 25 µL containing 10 ng of template DNA, 20 µL premix (Bioneer Co., Daejeon, Korea) and the 1.0 unit primer. Amplification products were separated by 1.4% agarose (Bioneer Co., Daejeon, Korea) gel electrophoresis with TBE (90 mM Tris, pH 8.5; 90 mM borate; 2.5 mM EDTA). The 100 bp DNA Ladder (Bioneer Co., Daejeon, Korea) was used as DNA molecular weight marker. The agarose gels electrophoresed were stained with ethidium bromide. The fragments were illuminated with ultraviolet ray and then photographed by photoman direct copy system (PECA products, Beloit, WI, USA).
Only the fragments ranged that were readily visible were scored for the statistical analysis. Bandsharing (BS) values were calculated according to the presence/absence of amplified products at specific positions in the same gel from the DNA profiles. Absence of fragments indicates that the priming site is not present, presumably because of some alteration in the DNA sequence. The degree of variability was calculated by use of the Dice coefficient (F), which is given by the formula: F=2 nab/(na+nb), where nab is the number of bands shared between the samples a and b, na is the total number of bands for sample a and nb is the total number of bands for sample b (Jeffreys & Morton, 1987; Yoke-Kqueen & Radu, 2006). The average of within-species similarity is calculated by the pairwise comparison between individuals within a species. The relatedness among different individuals of GRV and GRC of Jangheung generated according to the BS values and similarity matrix. The hierarchical clustering tree was analyzed by the similarity matrices to generate a dendrogram using pc-package program Systat version 10 (SPSS Inc., Chicago, IL, USA). The genetic difference and Euclidean genetic distance within- and between-species were calculated with hierarchical dendrogram program Systat version 10. All data were expressed as mean±SE. Significance was accepted for all tests at p<0.01.
RESULTS AND DISCUSSION
In spite of the variation in the DNA profiles and the difference in reproducibility, many genetic researchers used the DNA techniques because this PCR method is a relatively rapid, reliable and conveniently useful method to investigate numerous genomic DNAs for the genetic diversity in a population as well as it does not require the prior knowledge of the genome (Orozco-Castillo et al., 1994; Iyengar et al., 2000; Klinbunga et al., 2000a, Klinbunga et al., 2000b). The polymorphisms are determined by the banding patterns of amplified products at the specific positions by primers (Tassanakajon et al., 1998; Nozaki et al., 2000; Yoon & Kim, 2003b; Kim et al., 2004; Yoon & Kim, 2004; Song & Yoon, 2013). Thus, these DNA-based techniques have been applied to the identification of the genetic characteristic of diverse species of plants and/or seaweeds (Moeller & Schaal, 1999; Adams, 2000; Esselman et al., 2000; Kim & Choi, 2003; Kwon et al., 2004).
Here, the complexity of the banding patterns varied dramatically among the oligonucleotides primers from the two algal species. The sizes of the DNA fragments also varied wildly, from 90 to 2,400 bp, as shown in Fig. 1. Generally, the size and the number of the bands generated unbiased depends on the nucleotide sequence of the primer used and on the source of the template DNA, resulting in a genome-specific DNA band (Welsh & McClelland, 1990). When the plant universal primer set were used for PCR amplification in 18 genera and 26 species of seaweeds, a single or a few bands of 200–600 bp was generated (Hong et al., 1997). One to eight DNA bands were amplified, ranging from approximately 240 bp to 1,200 bp in seaweed Hizikia fusiformis (Park et al., 1998). The total amplified products from the six isolates were: 143 bands from the Chungmu sample, 135 bands from Haenam, 72 bands from Kijang, 120 bands from Pusan, 136 bands from Wando, and 109 bands from Yosu. In sorghum, 17 primers amplified from 4 to 12 DNA bands in the size range 500–2,500 bp (Jaiswal et al., 1998). In pumpkin species including C. maxima, C. moschata and C. pepo, a total of 136 RAPD markers were produced and 88 DNA bands were polymorphic with an average 5.5 polymorphic bands per primer (Kwon et al., 2004).
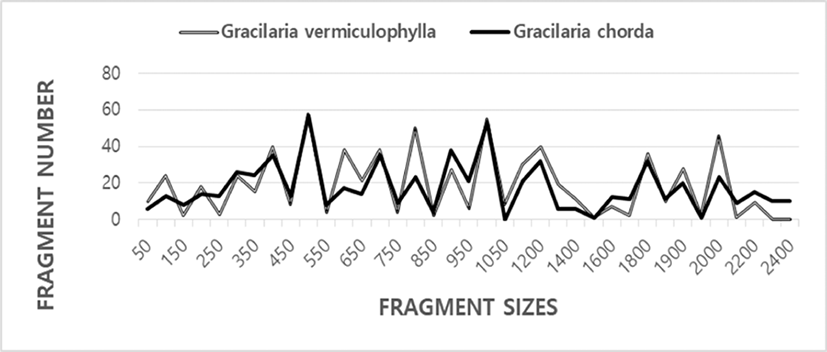
In the present study, the BS value based on the presence or absence of amplified bands was utilized to calculate the similarity indices in two Gracilaria species, as illustrated in Table 1. The similarity values based on the average BS value within GRV was 0.859±0.004, and of GRC was 0.916±0.006. BS value between two Gracilaria species ranged from 0.250 to 0.415 with the average 0.340±0.003. The value difference between the two Gracilaria species is statistically significant (p<0.01). Compared individuals searately, the BS value of individuals within GRC was higher than GRV species. Our BS values between two Graciaria species are similar to the result of Park et al. (1998) reported that the genetic similarity for the six isolates of seaweed Hizikia fusiformis ranged from 23% to 59%. Our BS values are also similar to previously reported results of Kim et al. (1997a) that similarity values obtained by RnRc primer analysis of nuclear DNA varied from 0.364 to 0.714 between Porphyra tenera (wild) and Porhyra tenera (Ariake).
Here, the number of unique shared loci to each species and number of shared loci by the two species generated by PCR using seven oligonucleotides primers in Gracilaria species from Jangheung, as demonstrated in Table 2. Two hundred and fifty-three unique shared loci to each species, with an average of 36.1 per primer, were observed in GRV species. Two hundred and forty-two unique shared loci, with an average of 34.6 per primer, were identified in the GRC from Jangheung. We have also identified 11 number of unique shared loci to each species in GRV, as revealed in Table 2. Eleven number of unique shared loci to each species generated by the oligonucleotides primer OPC-01 exhibited the inter-individual-specific characteristics and DNA polymorphisms, as illustrated in Table 2. Using numerous primers, PCR method has been applied to identify specific and/or polymorphic markers particular to line, species, genus and geographical population as well as genetic similarity/polymorphism/diversity in organisms (Callejas & Ochando, 1998; Klinbunga et al., 2000a; Yoon & Kim, 2003b; Yoon & Kim, 2004; Song & Yoon, 2013; Oh & Yoon, 2014). In the case of seaweed Hizikia fusiformis, thirty-one primers resulted in 715 PCR products of conserved and polymorphic bands (Park et al., 1998). Seventy primers produced 104 polymorphic bands, of which 89 were assigned to linkage groups in lentil (Lens sp.) (Eujayl et al., 1998). A genetic linkage map of Lens sp. was constructed with 89 RAPD markers, 79 AFLP markers and six RFLP markers. 38 primers were selected for analysis of total DNA RAPD and resulted in a total of 254 DNA fragments, of which 65% were polymorphic in sorghum (Jaiswal et al., 1998). The number of amplified bands generated by a primer varied from 19 to 27. 98.9% of these bands were polymorphic in Digitalis (Nebauer et al., 2000).
132 numbers of the shared loci by the two species, with an average of 18.9 per primer, were observed in the two Gracilaria species, as displayed in Table 2. The oligonucleotide primer OPA-04, OPD-08 and OPB-10 also generated 22 number of the shared loci by the two species, respectively. Particularly, 66 number of shared loci by the two species generated by the primer OPD-07 were identified commonly in two Gracilaria species, which identified populations and/or species. The specific primer was found to be useful in the identification of individuals and/or population, resulting from the different DNA polymorphism among individuals/species/population (Yoon & Park, 2002; Yoon & Kim, 2003a; Song & Yoon, 2013; Oh & Yoon, 2014; Yoon, 2015). Although the main disadvantage of this PCR method is its reproducibility, the method was considered suitable for the identification of a species. This technique was used to characterize six isolates of Hizikia fusiformis collected from six locales in Korea (Park et al., 1998). Representative isolate-specific and polymorphic banding patterns observed among the six isolates with specific RAPD primer. 3 out of 40 nanomer primers that amplified DNA bands specific to a grass species or to a durum cultivar were identified (Bommineni et al., 1997). The primer PR21 amplified DNA bands specific to five individual durum cultivars. The primers PR 22 amplified bands specific to a grass species. Similarly, the primer PR23 amplified bands specific to a grass species. The amplified bands ranging from 1,000 to 1,200 bp were specific to Thinopyrum junceiforme. The DNA band approximately 1.5kb was specificity to Lophopyrum elongatum. Generally, polymorphic loci generated by PCR using oligonucleotides primers were suitable to detect genetic similarity/diversity/ polymorphism among various organisms (Bommineni et al., 1997; Tassanakajon et al., 1998; McCormack et al., 2000; Nicolosi et al., 2000; Yoon & Kim, 2003a; Kim et al., 2004; Oh & Yoon, 2014; Yoon, 2015).
Based on the similarity matrix generated by BS values and genetic distances, hierarchical clustering analysis was performed to obtain the polar dendrogram, as shown in Fig. 2. The polar dendrogram obtained by the seven oligonucleotides primers, indicates two genetic clusters. The geetic distance between two Gracilaria species ranged from 0.059 to 0.513. The individual VERMICULOPHYLLA no. 07 of GRV was genetically closely related to VERMICULOPHYLLA no. 06 of GRV (genetic distance=.059). Especially, two entities between the individual VERMICULOPHYLLA no. 10 of GRV and CHORDA no. 22 of GRC showed the longest genetic distance (0.513) in comparison with other individuals used, as illustrated in Fig. 2. Our cluster analysis showed the similar pattern illustrated by Yoon and Kim (2004) that the single linkage cluster analysis indicating four genetic groupings and dendrogram revealed close relationships between individual identities within two geographical populations of Korean catfish (S. asotus) and bullhead (P. fulvidraco). In cyanobacterial picoplankton strains, the sequences of the three strains from Lake Biwa, Japan were closely related to those of marine cyanobacterial picoplankton strains and Prochlorococcus marinus found in the Pacific and Atlantic Ocean (Kane et al., 1997). In plant, RAPD data analysis of genetic distance and parsimony methods, family clustering, and the analysis of molecular variance were applied to study genetic relationships of a few of species within a genus. They reported that the species relationships revealed by RAPD-PCR approach should be consistent with the previously obtained data using morphological affinities (Nebauer et al., 2000). In this dendrogram, the 42 individuals grouped into seven main clusters, as would be expected from their morphological characters. The pattern of our cluster analysis was similar to that described by Orozco-Castillo et al. (1994). They reported that PCR analysis reflected morphological differences between the sub-groups as well as the morphological origin of the coffee plants.
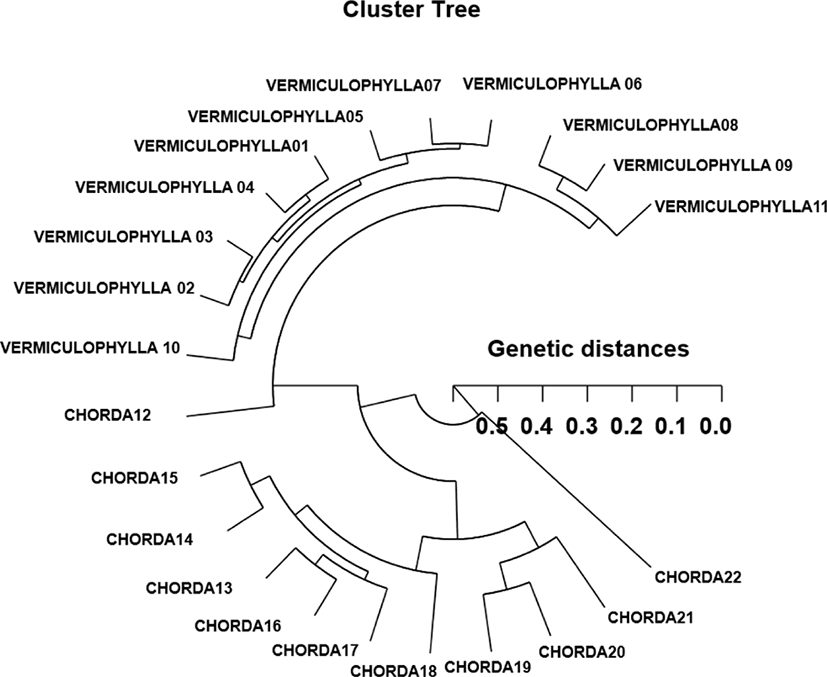
In our study, this PCR analysis has revealed a significant genetic distance between two Gracilaria species pairs (p<0.01). This shows that the method is an adequate tool to compare DNA in individuals, species and populations. Furthermore, the basic knowledge of DNA polymorphisms and molecular markers of Gracilaria species may contribute significantly to the seedling selection and the selective seaweed-breeding program. The classification of geographical populations and/or species of Gracilaria needs to be based on the morphological variations of seaweed blades such as size, shape, type, and color. As mentioned above, the potential of this PCR technique to identify diagnostic markers for strain, breed, species, genus, population and family identification in vegetation and/or seaweeds (Kane et al., 1997; Kim et al., 1997a; Eujayl et al., 1998; Moeller & Schaal, 1999; Adams, 2000; Bartish et al., 2000; Esselman et al., 2000; Nozaki et al., 2000; Kim & Choi, 2003; Kwon et al., 2004) has also been demonstrated.

