INTRODUCTION
Fish in the Tetraodontidae family are found in both freshwater and brackish waters of tropical and temperate coastal areas. Although many species in this family are economically important fishery resources, with unique flavors and high market value, they are often difficult to classify due to their wide variety of morphological variation (Kim & Lee, 1990; Tyler, 1980). Within Tetraodontidae, most pufferfish species in the genus Takifugu are highly toxic to humans, containing tetrodotoxins that affect a variety of organs including the liver and ovaries. Toxicity varies considerably among regions and seasons; therefore, inaccurate Takifugu species classification carries a risk of tetrodotoxin poisoning. The Ministry of Food and Drug Safety of Korea allows the trade of only 21 domestic and imported pufferfish species; however, some of these species have very similar morphologies and are difficult to distinguish. Trade disputes arising from problems with species identification and classification among six Takifugu species commonly consumed in Korea have motivated the search for a simple, rapid, and effective molecular genetic method for identification of these species.
Previous studies have applied morphological and component analyses to determine the species used in fishery products; recently, diagnostic polymorphic sites have been used to identify closely related species (Unseld et al., 1995; Kim et al., 2014). Molecular identification assays based on mtDNA sequences have been widely used to identify the origins of fishery resources and products for the prevention of fraud, mislabeling, and health risks (Rasmussen & Morrissey, 2008; Cohen et al., 2009; Acar et al., 2017). However, little effort has been devoted to molecular-based species identification of Takifugu pufferfish species (Hsieh & Hwang, 2004; Ishizaki et al., 2006; Hsieh et al., 2010; Luekasemsuk et al., 2015).
We developed a Takifugu species identification method that will contribute to the establishment of a safe fishery management and distribution system. We designed species-specific primers in the mtDNA cytochrome c oxidase subunit I (COI) gene region for the rapid and accurate identification of six Takifugu species, all of which are consumed in Korea: Takifugu pardalis, T. porphyreus, T. niphobles, T. poecilonotus, T. rubripes, and T. xanthpterus.
MATERIALS AND METHODS
For genetic analyses, we collected muscle tissue samples from T. pardalis (n=8), T. porphyreus (n=28), T. niphobles (n=8), T. poecilonotus (n=6), T. rubripes (n=23), and T. xanthpterus (n=198) stored by the National Institute of Fisheries Science (NIFS) and/or Marine Fish Resource Bank of Korea (MFRBK), and identified their morphological characteristics. These samples were collected in sterile tubes and preserved in 99.99% ethanol until DNA extraction. Total DNA from each sample was extracted using an automated DNA extraction system (MagExtractor MFX-6100, Toyobo, Osaka, Japan). Genomic DNA was quantified using a spectrophotometer (Nanodrop ND-1000, Thermo Fisher Scientific, USA) and stored at −20°C until genetic analyses.
We analyzed complete sequences of the mtDNA COI gene registered with the National Center for Biotechnology Information (NCBI) for the six pufferfish species: T. pardalis (GenBank accession no., AP009528.1), T. porphyreus (KY514076.1), T. niphobles (KY514069.1), T. poecilonotus (AP009539.1), T. rubripes (KP641572.1), and T. xanthpterus (KP641579.1) using the BioEdit v. 7.0.0 software to identify inter- and intra-species variation and conserved regions. Then we designed common primers (Taki-F35, Taki-R1232) specific to the six species (Fig. 1A). Polymerase chain reaction (PCR) was performed in a total volume of 20 μL, comprising 2 μL genomic DNA (20 ng), 0.6 μL dNTP (250 μM), 2 μL 1× PCR buffer containing 2 mM MgCl2, 0.4μL 10 pmol forward primer, 0.4 μL 10 pmol reverse primer, 0.2 μL 0.5 U DNA Taq (Anti-HS Taq, TNT Research, Seoul, Korea). The primers used are listed in Table 1. PCR amplification was performed using an ABI 2720 Thermal Cycler under the following conditions: 10 min of initial denaturation at 95°C; 37 cycles of 45 s at 94°C, 45 s at 58°C, and 1 min at 72°C; and a final extension for 5 min at 72°C. Amplified PCR products were sequenced using an ABI BigDye Terminator Cycle Sequencing Kit (ver. 3.1, Applied Biosystems, Foster, CA, USA), and analyzed using an ABI 3730XL DNA analyzer (Applied Biosystems).
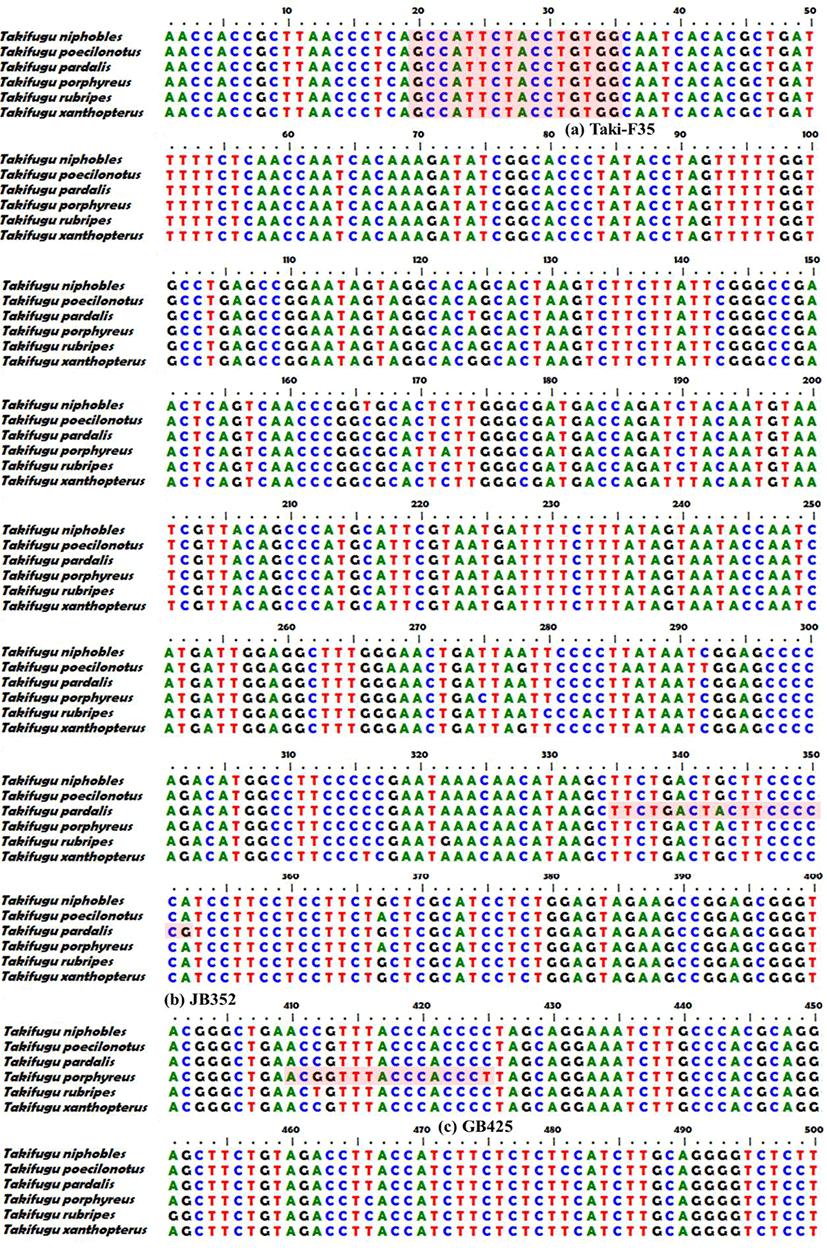
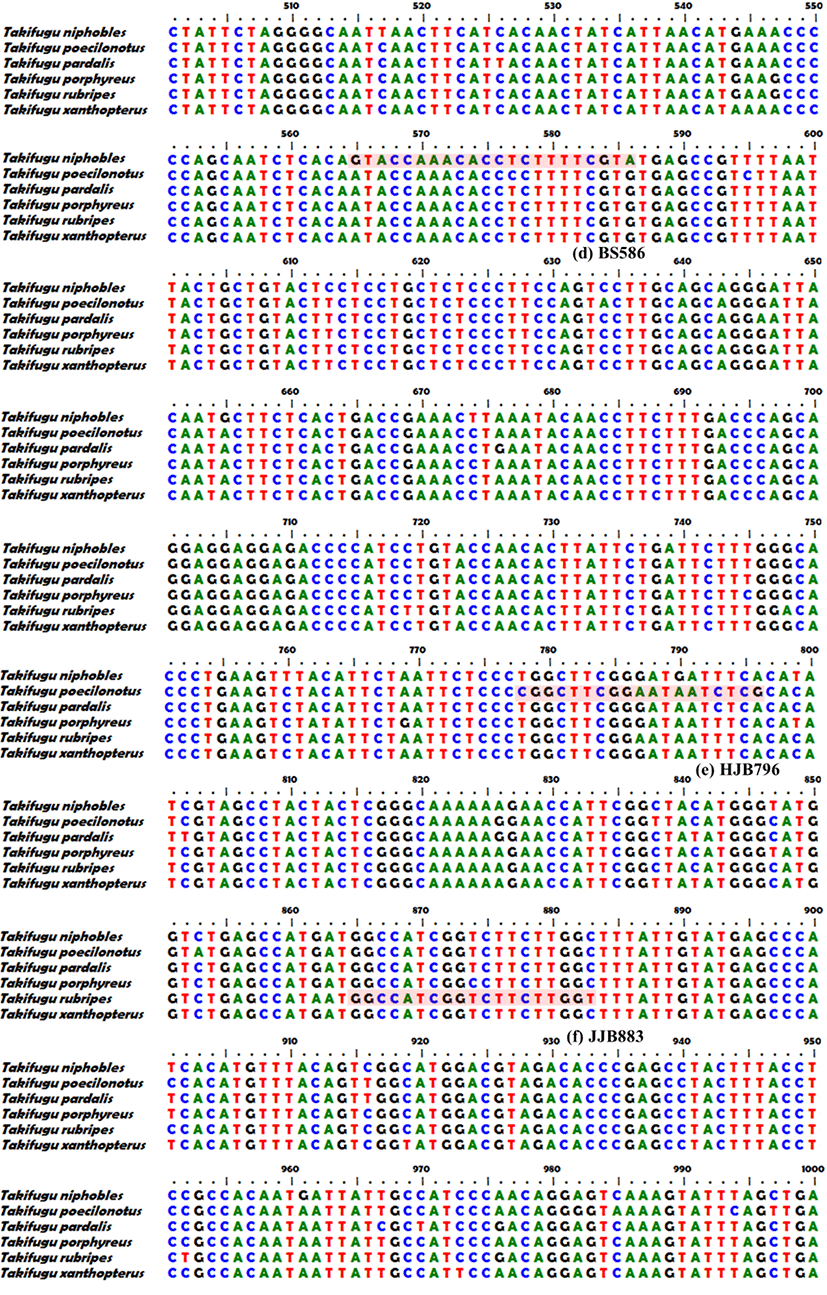
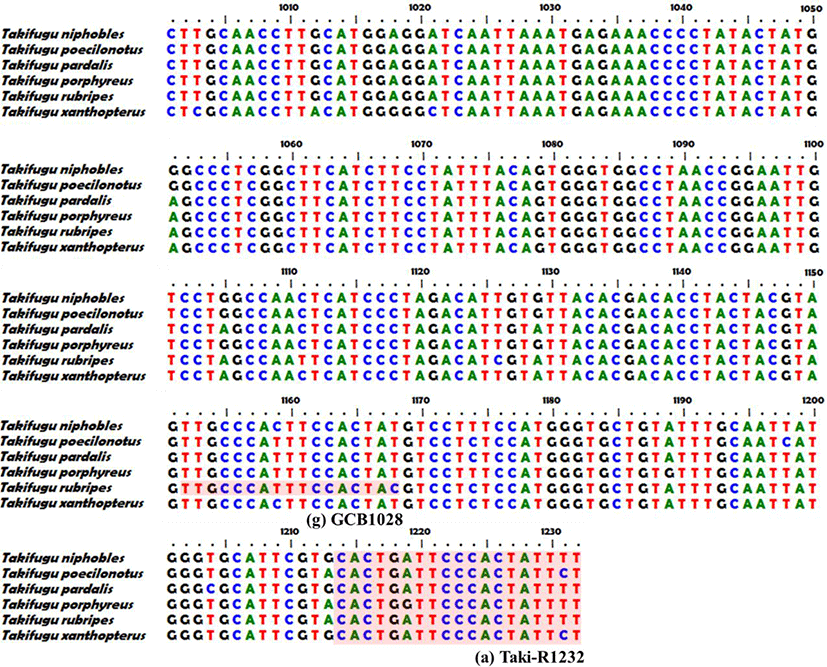
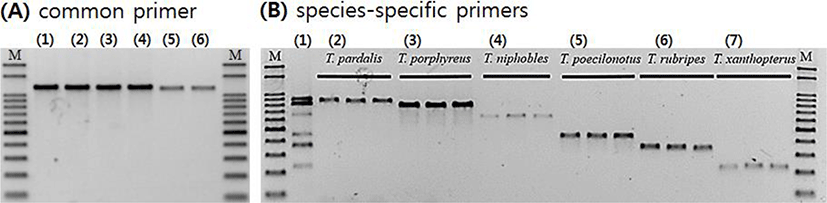
We assembled forward and reverse sequences of the six Takifugu species with common primers using the SeqMan software (DNASTAR, USA), and searched for single nucleotide polymorphisms (SNPs) showing species specificity except for intraspecies point mutation. Then we designed species-specific forward primers for each target species in which the SNP was located at the 3’ end (Fig. 2B(1)–(7), Table 1). We performed species-specific PCR analyses using the six forward primers (JB352, GB425, BS586, HJB796, JJB883, and GCB1028) and one reverse primer (Taki-R1232) under the PCR conditions described above for the common primer, but at an annealing temperature of 60°C. Amplified PCR products were subjected to 2% agarose gel electrophoresis (100 V, 40 min) with 1× RedSafe (iNtRon, Korea), and confirmed using the Gel Doc image analysis system (ATTO Corp., Japan).
RESULTS AND DISCUSSION
A region of the cytochrome COI is widely applied in genetic identification of animals including fishes and improved effectiveness in species identification on the previous report (Rasmussen & Morrissey, 2008; Cohen et al., 2009; Acar et al., 2017). PCR analyses using species-specific primers have excellent sensitivity and specificity, and can analyze large numbers of samples within a short time. These methods are widely used for species identification because they are simpler than DNA sequence analyses (Hwang et al., 2002). Also, MSS-PCR methods are economical in terms of time, effort, and cost compared to other DNA based methods, and the determination of the most important species can provide clear and repeatable results (Sezaki et al., 2005; Noh et al., 2017).
We successfully developed and applied a multiplex PCR assay based on species-specific variation for rapid and simultaneous identification of six target species: T. pardalis, T. porphyreus, T. niphobles, T. poecilonotus, T. rubripes, and T. xanthpterus. This is the first study to use conventional PCR amplification to identify six pufferfish species commonly imported into the Korean fishery market.
We obtained 1,252 bp sequences of the mtDNA COI region from the six species, and used a 1,197 bp common primer to confirm MSS-PCR amplification (Fig. 1). Sequences obtained using the common primer were analyzed using the DNA Sequence Polymorphism (DnaSP) v. 5.10.01 software. Haplotype analysis results for a total of 271 individuals among the six species indicated 4 haplotypes of T. pardalis (n=8), 4 of T. porphyreus (n=28), 7 of T. niphobles (n=8), 4 of T. poecilonotus (n=6), 6 of T. rubripes (n=23), and 32 of T. xanthpterus (n=198). Except for intraspecies genetic variation, species-specific SNPs were observed at 352 bp (JB352), 425 bp (GB425), 586 bp (BS586), 796 bp (HJB796), 883 bp (JJB883), and 1,028 bp (GCB1028) (Fig. 1).
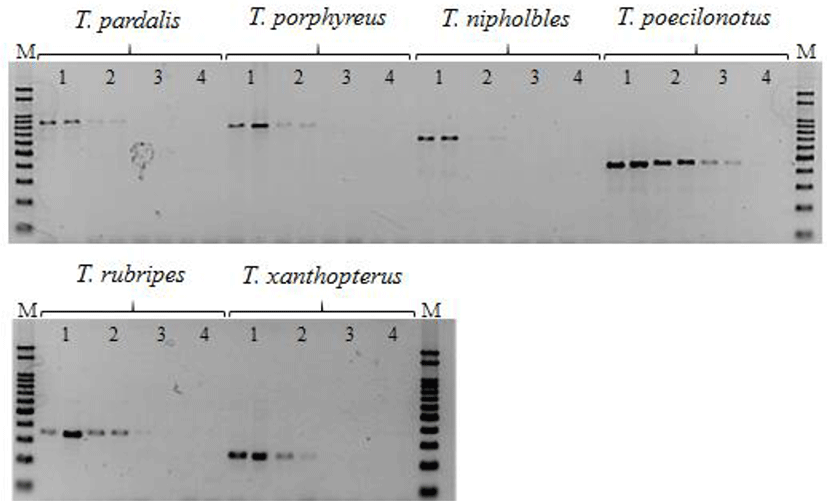
MSS-PCR products containing the same amounts of forward primers were confirmed by agarose gel electrophoresis, with accurate DNA amplification of each species and clear distinction among amplified products by size (Table 1). Sequencing analyses confirmed that MSS-PCR products of the six species had 100% identity with the expected regions. Primer dimers and nonspecific amplification products were not observed, and no cross-reactions were observed among species-specific amplifications in DNA mixtures of the six species (Fig. 2). The MSS-PCR assay sensitivity test showed that DNA template concentrations of stored products from the six species were 10 ng/μL, 1 ng/μL, 0.1 ng/μL, and 0.01 ng/μL. The PCR assay sensitivity of each species was detectable to a concentration of 1 ng/μL among all six species (Fig. 3).
In conclusion, the molecular method developed in this study was simple, rapid, and inexpensive compared to direct sequencing analyses, and did not require high-quality equipment. The MSS-PCR method is capable of clearly distinguishing and/or authenticating six Takifugu species in case of mislabeling via accident or fraud. This technique can be used as a regulatory tool to protect public health and enforce Korean fishery product import regulations.

