INTRODUCTION
Neural precursors (NPs) derived from human embryonic stem cells (hESCs) are highly proliferative and can give rise to three types of cells of the central nervous system (CNS): neurons, astrocytes, and oligodendrocytes. The primitive nature of NPs enables them to be engineered to form specific neural cells found in particular regions of the CNS, such as spinal motor neurons and midbrain dopaminergic neurons (Kriks et al., 2011; Du et al., 2015); this is a unique property of NPs derived from hESCs that is nearly impossible to see in neural stem cells from fetal and adult tissues. In this regard, hESC- derived NPs are an invaluable source for biomedical applications.
Technologies for the derivation of NPs from hESCs have been intensively developed. Based on their experience with mouse ESC culture, scientists have developed several methods for neural induction of hESCs: 1) the embryoid body (EB)-based method, in which EBs made using a suspension culture of hESCs are attached to generate a “neural rosette,” a characteristic morphology of primitive NPs, under neural tissue-inducing culture conditions (Zhang et al., 2001) and 2) coculture of hESCs with mouse stromal cell lines, such as PA6 and MS5, which is often employed to generate neural rosettes and NPs from hESCs (Perrier et al., 2004). More recently, a great advancement has been achieved by adopting the concept of neural development in vertebrate embryos. The simultaneous inhibition of cellular signals that suppress neuroectodermal development – bone morphogenetic protein (BMP) and transforming growth factor-β (TGF-β) signals – could robustly enhance the efficiency of neural differentiation from hESCs (Chambers et al., 2009). This strategy, often referred to as “dual-SMAD inhibition,” has even helped overcome the differentiation propensity observed in many hESC lines that have proven stubborn to differentiate into cells of neural lineage (Kim et al., 2010). Due to the robustness of the results obtained using it, this method has now become the most popular protocol for generating NPs from hESCs.
While the involvement of cell signaling pathways, such as BMP and TGF-β pathways, in neural induction has been intensively studied for decades, the contribution of the extracellular environment to neural induction has just begun to attract the attention of stem cell scientists. The extracellular matrix (ECM) greatly influences the survival, proliferation, and differentiation of stem cells (Niklason, 2018). For instance, specific ECM molecules, such as laminin and fibronectin, influence neural differentiation and neurite outgrowth through cellular signals mediated by integrin receptors (α6β1 or α5β1). In addition to such roles, the physical properties of ECMs, such as stiffness and elasticity, have also been discovered to be important for differentiation of hESCs toward a neural lineage (Akhmanova et al., 2015) and determination of the fate of specific neural cells (Smith et al., 2018). More recently, synthetic nanofibers, inspired by the nano-architecture of natural ECM, have been shown to enhance the neural differentiation of hESCs (KarbalaeiMahdi et al., 2017), indicating the role of extracellular environment in neural lineage specification.
In this study, we sought to develop a new platform for the neural differentiation of hESCs by creating a nanofiber scaffold composed of a biocompatible and electroactive material, polyvinylidene fluoride (PVDF). In particular, we adopted an electrospinning technology to produce a scaffold of nanofibers with a defined diameter and immobilized a vitronectin-derived peptide on the scaffold to maximize the adherence of hESCs. When hESCs were made to differentiate under dual-SMAD inhibition, we found that the nanofiber scaffold facilitated neural differentiation more efficiently than Matrigel, a conventional ECM, and the difference in efficiency was statistically significant. RNA sequencing (RNAseq) analysis revealed several key molecular changes relevant to the neural development of hESCs grown on our PVDF nanofiber scaffold. These results substantiate the utility of the PVDF nanofiber scaffold for the differentiation of NPs from hESCs and its potential as a new platform for future biomedical applications.
MATERIAlS AND METHODS
Preparation of the PVDF nanofiber scaffolds was performed by AMO Greentech (Gimpo, Gyuonggi-do, Korea) and AMO Lifescience (Seoul, Korea) using a spin-coated PVDF solution dissolved in a dimethylacetamide solvent (Sigma, St Louis, MO, USA). Briefly, the spinning solution was loaded in a syringe capped with a nozzle tip whose outer diameter was 0.9 mm; it was then electro-sprayed at a power of 20–40 kV and a flow rate of 50–100 μg/mL. The nanofibers were compressed using lamination, with heat of 80°C–120°C. After drying at room temperature (RT), the compressed nanofiber scaffold was punched out in a circular pattern and fixed to the bottom of a 6-well plate using a polyethylene terephthalate (PET) film.
Mussel adhesive protein (MAP) is a natural glue that is derived from marine mussel and is often used for cell adhesion (Lee et al., 2011). We first cloned a plasmid with a gene encoding a recombinant MAP fusion protein composed of fp-1, as described elsewhere (Hwang et al., 2007). Then, based on the published amino acid and cDNA sequences available on the NCBI website (Gene ID: 7448), we cloned a plasmid encoding a fusion protein of MAP and a heparin-binding peptide of vitronectin that has been shown to support the attachment of hESCs (herein, VNm) (Klim et al., 2010). The recombinant hybrid MAP-VNm fusion protein was expressed in E. coli (Rosetta-gami®) (Thermo Fisher Scientific, Waltham, MA, USA) using a 14 L fermentor and harvested as inclusion bodies. Next, the inclusion bodies were recovered as a solution of functional peptides in distilled water by a conventional procedure (Palmer & Wingfield, 2004).
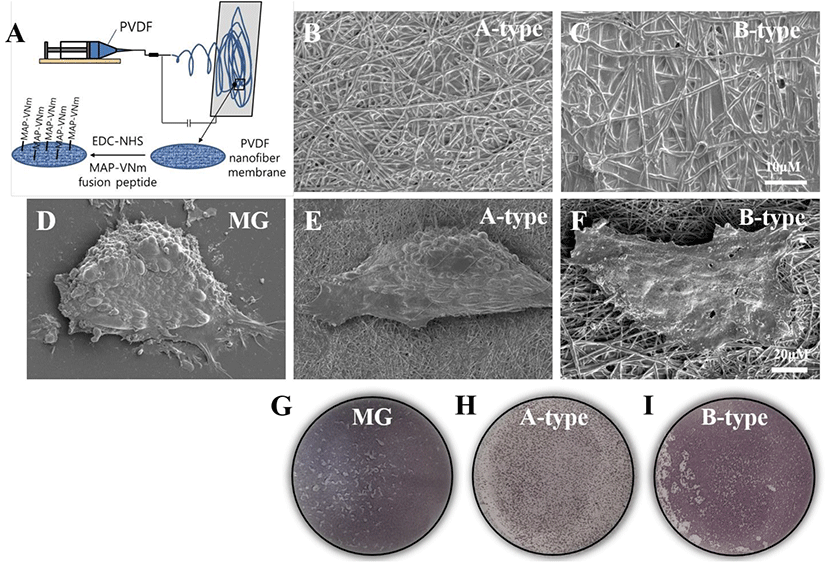
Before coating its surface with the recombinant MAP-VNm fusion protein, we activated the nanofiber scaffold with a 10 mM solution of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) (AK Scientific, Union City, CA, USA) and 10 mM N-hydroxysuccinimide (NHS) (AK Scientific) in 20 mM sodium acetate buffer (pH 6.5) at RT for 30 min. We then added a solution of MAP-VNm fusion protein (0.05 μg/mL) to the activated scaffold for 30 min at RT. After three washes with distilled water for 10 min, the scaffold coated with the fusion protein was dried and stored at RT before use. The schematic processes of scaffold fabrication and surface coating are illustrated in Fig. 1A.
An hESC line, WA09 (conventionally known as H9), was obtained from WiCell (USA). The hESCs were cultured on a Matrigel® (Thermo Fisher Scientific)-coated 6-well plate in StemMACS®iPS-Brew XF medium (Miltenyi Biotec, San Diego, CA, USA) (herein, hESC culture medium) and expanded by enzymatic passaging, a conventional expansion technique. For neural differentiation, we employed the dual-SMAD inhibition strategy previously described by Chambers et al. (Chambers et al., 2009), with slight modifications. Briefly, we plated hESCs on Matrigel or nanofiber scaffolds at the density of 1×104 cell/cm2 in hESC culture medium containing 10 μM Y-27632 (Sigma). From the day 1 of differentiation, we began to add 250 nM of LDN193189 (StemCell technology, Vancouver, BC, Canada) and 10 μM of SB431542 (Sigma) in the hESC culture medium and cultured the hESCs for 8 days. On day 4, we switched the culture medium from hESC culture medium to neural induction medium (DMEM/F12 medium supplemented with 1× N2, 1× non-essential amino acid (all from Thermo Fisher Scientific) and 20 ng/mL bFGF (Peprotech, Rocky Hill, NJ, USA)). The medium was replenished on alternate days. On day 8, the cells were either harvested for RNA preparation or dissociated by Accutase (Thermo Fisher Scientific) and replated on Matrigel-coated 12 mm coverslips for immunostaining. For further differentiation, on day 8, the cells were dissociated and replated on a Matrigel-coated coverslip at the density of 5×104 cell/cm2. The cells (NPs) were induced to differentiate in neural differentiation medium (DMEM/F12 supplemented with 1× N2, 1× B27, 1× non-essential amino acid, as well as 10 ng/mL BDNF (Peprotech) and 20 μM ascorbic acid (Sigma)) for 2 weeks.
Cell samples were fixed in 4% paraformaldehyde for 15 min at RT and washed with phosphate buffed saline (PBS). After permeabilization with 0.05% Triton X-100 in PBS for 5 min, the samples were blocked using 2% bovine serum albumin solution for at least one hour and then incubated with primary antibodies (see below) overnight at 4°C. After that, the samples were thoroughly washed with PBS thrice and then incubated with appropriate secondary antibodies conjugated with a fluorescent dye (Alexa fluor® 488 or 594, Thermo Fisher Scientific) for 30 min at RT. After washing with PBS, the coverslips were mounted on glass slides using a 4’,6-diamidino- 2-phenylindole (DAPI)-containing mounting solution (Vector laboratory, Burlingame, CA, USA). The cell images were captured using a fluorescence microscope (IX71) equipped with a digital camera (DP71) (both from Olympus, Tokyo, Japan). Primary antibodies used in this study were as follows: SOX1 (rabbit, 1:200; R&D Systems, San Diego, CA, USA), SOX2 (rabbit, 1:200; Thermo Fisher Scientific), NESTIN (Mouse, 1:200; Thermo Fisher Scientific), TUJ1 (Mouse, 1:1,000; BioLegend, San Diego, CA, USA), GFAP (Rabbit, 1:1,000; Dako, Carpinteria, CA, USA), A2B5 (Mouse, 1:200; Thermo Fisher Scientific), and NG2 (Rabbit, 1:100; Thermo Fisher Scientific).
Total RNAs were extracted using the Easy-Spin® total RNA purification kit (iNtRON Biotechnology, Seongnam, Gyeonggi-do, Korea) according to the manufacturer’s instruction. One microgram of the total RNAs was used to synthesize cDNAs using the Power cDNA Synthesis kit (iNtRON Biotechnology). Next, qRT-PCR was performed on the StepOnePlus® real-time PCR system (Thermo Fisher Scientific) using the Fast SYBR® Green Master Mix (Thermo Fisher Scientific). Primer sequences are listed in Table 1.
The cell specimens were fixed with 2.5% glutaraldehyde solution and incubated at 4°C overnight followed by washing with 0.1 M cacodylate buffer (pH 7.2). The cells were then incubated in 1% osmium tetroxide for 1 hour, washed with distilled water, and dehydrated gradually using increasing concentrations of ethanol (50%, 70%, 90%, and 100%). After drying inside a fume hood, the cell specimens were mounted on SEM stubs followed by platinum coating with a sputter coater (E-1045, Hitachi, Tokyo, Japan). Surface imaging was carried out using the ETD detector of an SEM (TeneoVS, FEI) in the SE mode at 2 kV.
Total RNA was isolated as previously described and was submitted to MACROGEN, Korea for paired-end sequencing; over 60 million reads were generated. Raw FASTQ files were trimmed for adapters using TrimGalore (Krueger, 2020), quantified at the transcript level against an Ensembl catalog using Salmon (Patro et al., 2017), aggregated to the gene level using tximport (Soneson et al., 2015), and delivered to DESeq2 (Love et al., 2014) for further analysis using a standard pipeline. A correlation plot was prepared using ggplot2 (v3.3.0), and a heatmap was prepared using pheatmap (v1.0.12). Gene ontology analysis was performed using Metascape (Zhou et al., 2019).
The experiments were performed in triplicate at the least, and data were presented as mean±SEM. Experimental data was analyzed with one-way analysis of variance (ANOVA) followed by Bonferroni’s post hoc test. p<0.05 was considered to be statistically significant.
RESUlTS
To examine the behavior of hESCs grown on the PVDF nanofiber scaffolds, we first created two types of PVDF membranes with fiber diameters of 200 nm (referred to as A-type) and 700 nm (B-type), respectively. Our preliminary observation revealed that the electrospun PVDF nanofiber scaffolds were not compatible for the attachment of hESCs possibly due to their hydrophobic nature (data not shown). Therefore, to endow the PVDF nanofiber scaffolds with the ability to support the attachment of hESCs, we made a biochemical modification in the scaffolds by immobilizing a peptide motif derived from vitronectin (VNm) that was shown to support the attachment and growth of hESCs (Klim et al., 2010). In particular, we immobilized the VNm on the surface of the scaffold by creating a fusion peptide with MAP and combining it using a recent technology that involves a peptide-acrylate surface coating process (Fig. 1A) (Hwang et al., 2007; Melkoumian et al., 2010) (see the details in Materials and Methods). As shown in Fig. 1B and C, EM showed that each PVDF nanofiber scaffold was composed of nano-sized fibers with a relatively constant diameter and without any cobble-stone-like aggregation. The hESCs seeded and cultured on each scaffold were observed to be as adherent as on Matrigel (Fig. 1D–F), indicating the functional immobilization of the VNm peptide. Interestingly, the shape of hESC colonies on the A-type scaffold appeared similar to the shape of colonies on Matrigel, which is dome-shape, whereas the colonies of hESCs on the B-type scaffold were spread out and had a flat-shape (Fig. 1D–F). As PVDF nanofiber scaffolds are opaque, bright-field microscopy could not be used to observe the morphology of the cells; thus, AP staining was performed, and the morphology of cells was observed by EM.
To test whether the PVDF nanofiber scaffolds facilitate the neural differentiation of hESCs, we cultured hESCs on either nanofiber scaffolds or Matrigel (as a control ECM) under dual-SMAD inhibition and compared the efficiency of neural differentiation. On day 8 of differentiation, we detached the cells from the scaffolds and Matrigel by enzymatic dissociation, and replated them on a Matrigel-coated coverslip for immunocytochemistry. Immunostaining with an antibody against SOX1, a definitive marker for NPs, revealed that 81.3±0.3% of the cells in the Matrigel group had strong immunoreactivity (Fig. 2A and G), which is similar to the results of previous studies (Chambers et al., 2009). In contrast, we observed that of the total cells on the A-type and B-type scaffolds, 91±2.5% and 86±1.2% were positive for SOX1, respectively (Fig. 2B, C, and G). More importantly, the yield of SOX1-positive cells on both the nanofiber scaffolds was significantly higher than that on Matrigel (Fig. 2G) (p=0.008 for A-type and p=0.04 for B-type, n=3 in each group, one-way ANOVA test with Bonferroni’s post hoc test). In addition, almost all cells were also positive for SOX2 and NESTIN, ensuring their fate as NPs (Fig. 2D–F). Relative gene expression analysis for neural precursor markers, such as NESTIN and PLZF, also revealed the significantly (in case of NESTIN) increased expression of the markers in the nanofiber scaffold groups compared to that in the Matrigel group (Fig. 2H and I). Collectively, these results suggest that PVDF nanofiber scaffolds coated with VNm cause better neural differentiation of hESCs than the conventional ECM(Fig. 1G-I).
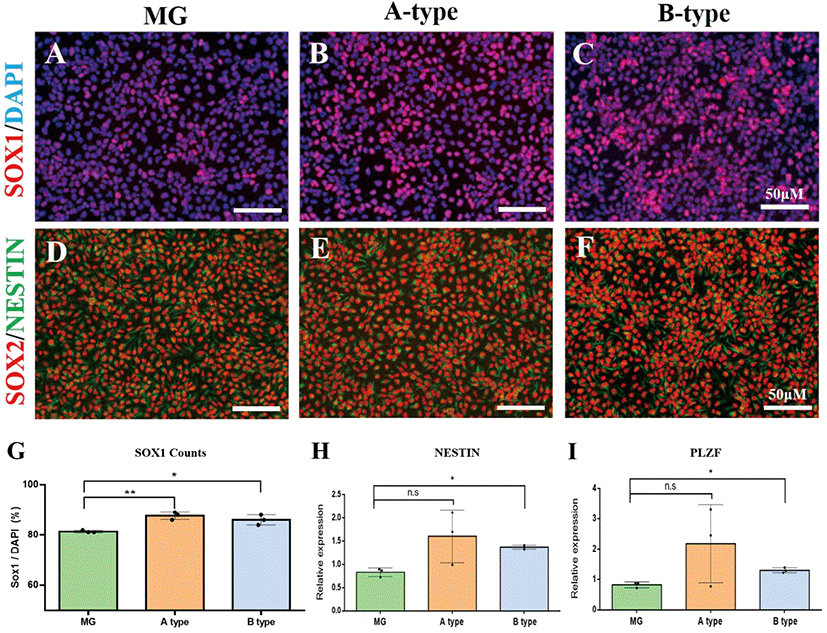
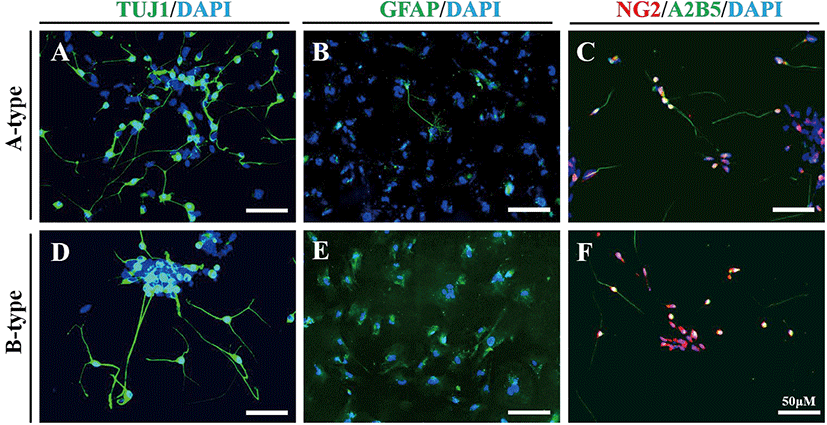
Next, we examined whether the NPs differentiated on nanofiber scaffolds retain their neural stem cell differentiation potential. We further differentiated NPs by culturing them in neural differentiation medium. In two weeks, NPs generated on the PVDF nanofiber scaffolds differentiated into TUJ1-positive neuronal cells, GFAP-positive glial cells, and NG2/A2B5-double positive oligodendrocyte precursor cells (Fig. 3). Of the NPs generated on the PVDF nanofiber scaffolds, TUJ1-positive cells were most frequently observed, followed by NG2/A2B5-double positive cells and GFAP-positive cells. Thus, we concluded that NPs differentiated on the PVDF nanofiber scaffolds retain their differentiation potential as neural stem cells.
Next, we performed comparative transcriptome analysis by RNAseq to explore the mechanism through which the PVDF nanofiber scaffold facilitated neural differentiation of hESCs. Hierarchical clustering showed that the two nanofiber scaffold groups were more closely related to each other than the Matrigel group (Fig. 4A); this was consistent with the results regarding the yield of SOX1-positive cells (Fig. 2D). We identified 45 genes with highest variation in expression across samples. Of those, 6 genes were upregulated exclusively in the Matrigel group and, among these, 3 genes had known gene annotations (in the green box of Fig. 4A and C; NELL2, CA4, and GRIN2B). In other words, NELL2, CA4, and GRIN2B were specifically downregulated in both the nanofiber scaffold groups. These genes play roles in protein kinase signaling, bicarbonate transport, and transportation of Ca2+ ions (Fig. 4C). In contrast, 27 genes were upregulated in the nanofiber scaffold groups, of which three and seven genes were exclusive to the A-type group and B-type group, respectively (Fig. 4B). Two out of three genes in the A-type group had known GO-terms related to the regulation of transcription (VGLL3) and FGF receptor signaling pathway (FGFBP3) (orange region in Fig. 4A and C). Given that FGF signaling is essential for neural induction during vertebrate development (Wilson & Edlund, 2001), upregulation of FGFBP3, a positive regulator of the FGF signal, might be the factor that enhances the efficiency of neural differentiation on the A-type nanofiber scaffold.
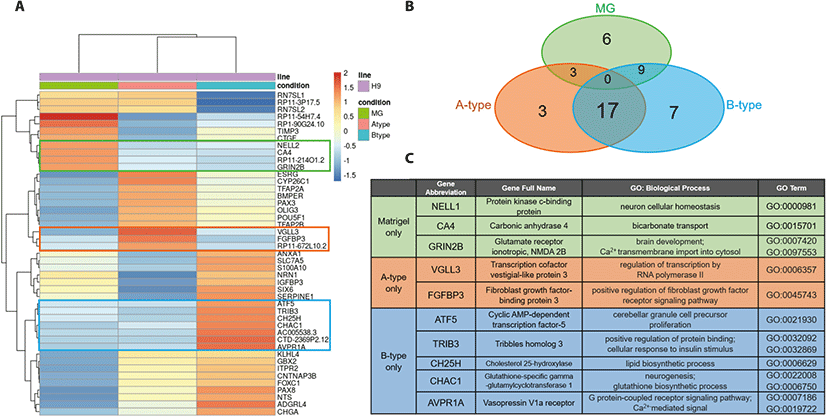
The gene-set specific for the B-type group included five genes (out of seven genes upregulated) that are related to neural cell proliferation (ATF5), cellular response to insulin (TRIB3), lipid metabolism (CH25H), glutathione biosynthesis (CHAC1), and G-protein-coupled receptor signaling pathway (AVPR1A) (blue region in Fig. 4C). The remaining 17 genes were shared by both the nanofiber scaffold groups (Fig. 4B, and Table 2). Among those, eight genes were expressed more highly in the A-type group than in the B-type group; the expression of the other nine genes was higher in the B-type group than in the A-type group (Table 2). A GO term search revealed that several of the identified genes are directly related to the development of the nervous system (e.g. CYP26C1, PAX3, OLIG3, GBX2, and PAX8). Other genes are known to be involved in biological processes, including development and morphogenesis (TFAP2A, POU5F1, and FOXC1), G protein-coupled receptor signaling (NTS and ADGRL4), and cellular signaling pathways that are relevant to neural induction (BMPER and TFAP2B). It is noteworthy that BMPER is upregulated in both the nanofiber scaffold groups. It has been a long-standing theory that the BMP signal inhibits neural induction (Wilson & Edlund, 2001) and that activation of the ERK signal is required for the acquisition of neural fate during early vertebrate development (Kuroda et al., 2005). Since BMPER is known to play dual roles in both BMP inhibition (Kelley et al., 2009) and ERK activation (Heinke et al., 2013), its upregulation can be considered as one of the key factors that facilitate neural differentiation on PVDF nanofiber scaffolds. Collectively, our finding that genes upregulated in the nanofiber scaffold groups are highly related to neural development suggests that these genes might play a significant role in the enhanced neural differentiation on PVDF nanofiber scaffolds.
ND, not determined; NA, not applicable; BMP, bone morphogenetic protein.
DISCUSSION
Dual-SMAD inhibition is to date the most efficient and powerful approach proposed for the neural differentiation of hESCs and forces hESCs to differentiate into cells of a neuroectodermal lineage without a choice to differentiate into cells of other lineages, such as endoderm, mesoderm, and even epidermal lineages (Chambers et al., 2009). This strategy was discovered and has been used on rigid and smooth plastic culture plates, yet it yielded a substantial percentage (~80% of total cells, see reference (Chambers et al., 2009) and Fig. 2G) of SOX1-positive NPs in 8 days of differentiation. This robustness might be the reason why no other strategy significantly improves the efficiency of neural differentiation in combination with dual-SMAD inhibition.
The physical cues from the fibrous architecture of the extracellular environment have been intensively explored in culture techniques for stem cells, particularly for the facilitation of differentiation efficiency (Li et al., 2014). In recent studies, researchers have produced nanofiber scaffolds with various biocompatible materials and showed that these scaffolds could replace the need of ECM substrates for neural differentiation of pluripotent stem cells (KarbalaeiMahdi et al., 2017; Silantyeva et al., 2018). These findings indicated the possibility that modulation of the extracellular environment may further increase the high efficiency of neural differentiation brought about by dual-SMAD inhibition.
In an attempt to devise a new synthetic microenvironment for the neural differentiation of hESCs, we fabricated a nanofiber scaffold with PVDF using the electrospinning technique. We were particularly interested in PVDF because of its unique piezoelectric property. Piezoelectricity is a quality of material asymmetry, which leads to the spontaneous generation of electric signals upon physical deformation (Li et al., 2019). Due to the unique arrangement of polar fluorine atoms within the vinylidene fluoride monomer (namely, β-phase), the electrospun nanofiber scaffold of PVDF is highly electroactive; thus, it has been used to culture or differentiate electrically active cells, such as cardiomyocytes and osteoblasts (Li et al., 2019). Based on the same concept, we hypothesized that PVDF nanofibers result in the formation of a scaffold that provides not only the fibrous microenvironment of brain tissue but also an electroactive substrate for the differentiation of neural cells from hESCs. Despite all its advantages, however, our previous observation indicated that the PVDF nanofiber scaffold is not compatible for the attachment of hESCs (unpublished). Therefore, we made a biochemical modification to PVDF for the adherence of hESCs by immobilizing a vitronectin peptide onto it through fusion with MAP combined with EDC/ NHS activation. As a result, hESCs were able to firmly adhere onto the nanofiber scaffolds. Most importantly, when hESCs were differentiated on the scaffolds under dual-SMAD inhibition, significantly more SOX1-positive NPs were generated on the scaffolds than on Matrigel. The NPs were able to differentiate into neurons, astrocytes, and oligodendrocyte precursors, indicating that the NPs completely retain their differentiation potential as neural stem cells.
Our comparative gene expression analysis revealed highly variable genes in the nanofiber scaffold groups. Among the 27 genes that were upregulated in the nanofiber scaffold groups, 17 genes were upregulated in both A- and B-type scaffolds, even though the extent of expression of individual genes differed slightly in each group. It was not surprising to see that majority of the genes (17/27 genes) were commonly upregulated in both the groups because both types of scaffolds were produced using the same technique (electrospinning) and the same material (PVDF). Nonetheless, we still found that 10 genes were exclusively upregulated in each scaffold group (3 genes in the A-type group and 7 genes in the B-type group). Because the only difference between the two scaffolds was the diameter of nanofiber, we infer that the topological effect of different diameters might have resulted in differential gene expression. In support of this idea, previous studies have shown that the fiber diameter of electrospun scaffolds influences differentiation of stem cells (Christopherson et al., 2009; Bean & Tuan, 2015). These studies proposed that change in membrane nanostructure could affect the protein binding properties of the scaffold, which could subsequently alter cell morphology and differentiation potential. As shown in Fig. 1E and F, the morphology of hESCs was indeed different on the two nanofiber scaffolds. Therefore, we assume that the morphological changes in the cells grown on nanofiber scaffolds with different fiber diameters altered the differentiation kinetics of hESCs and thus resulted in differential gene expression between the two groups.
In summary, this study provides a new culture platform for the neural differentiation of hESCs. The PVDF nanofiber scaffolds coated with a peptide derived from vitronectin efficiently supported the attachment of hESCs and, most importantly, were able to enhance the efficiency of neural differentiation using the dual-SMAD strategy. Since this new platform can be produced in a highly defined manner and is composed of a stable and biocompatible material, we believe that it will serve as an advanced culture system for the production of neural cells for future biomedical applications.

