INTRODUCTION
Female germline stem cells (GSCs) located in the anterior end of the Drosophila ovary continuously produce eggs throughout the life-span. In particular, GSCs attached to the somatic cap cells exhibit asymmetric cell division to generate a stem cell (self- renewal), which is also attached to the cap cells, and a cystoblast (CB), which is one cell away from the cap cells and enters a differentiation program (Spradling et al., 1997). The CB undergoes four incomplete divisions to generate a 16-cell cyst, one of which will be an oocyte and the others nurse cells.
Maintenance of GSCs is dependent on signals emanating from the cap cells, which construct a niche environment supporting the GSCs. Decapentaplegic (Dpp) is a critical extrinsic niche signal that stimulates continued stemness of GSCs (Xie & Spradling, 1998). In addition, intrinsic factors expressed in GSCs mediate stemness and block differentiation. The translational regulator Nanos (Nos) is one such intrinsic factor required for stemness maintenance (Lin & Spradling, 1997; Forbes & Lehmann, 1998). Nos collaborates with another translational regulator, Pumilio (Pum), to repress expression of factors that promote stem cell differentiation (Joly et al., 2013).
In contrast, Bag of marbles (Bam) is an intrinsic differentiation-promoting factor expressed in CBs (McKearin & Spradling, 1990). In a bam mutant ovary, GSCs fail to undergo differentiation and produce stem cell-like tumor cells (McKearin & Spradling, 1990). Upon ectopicexpression in GSCs, Bam promotes differentiation of stem cells (McKearin & Ohlstein, 1995; Ohlstein & McKearin, 1997). One role of Bam in differentiating cells is to repress Nos expression at the translational level through the nos 3’ untranslated region (UTR) (Li et al., 2009). Other critical differentiation-promoting factors include Benign gonial cell neoplasm (Bgcn), Sex lethal (Sxl), and Meiotic P26 (Mei-P26) (Chau et al., 2009; Ohlstein et al., 2000; Li et al., 2013). Bgcn encodes a protein distantly related to DExH-box RNA helicase, while Sxl is an RNA-binding protein with specificity to polyuridines stretches and Mei-P26 is an RNA-binding protein with an NHL (Ncl-1, HT2A and Lin-41) domain that functions as a translational repressor (Sekelsky et al., 1999; Glasscock et al., 2005; Chau et al., 2012). Genetic studies suggest that Bam functionality requires Bgcn, Sxl, and Mei-P26, suggesting that Bam acts in conjunction with these proteins (Chau et al., 2012; Li et al., 2013). However, the mechanics of how Bam recognizes nos mRNAs and how it collaborates with Bgcn, Sxl, and Mei-P26 remain poorly understood.
To understand the molecular mechanisms underlying Nos repression, we undertook rigorous protein-protein interaction studies and performed interaction mapping on Bam, Bgcn, Sxl, and Mei-P26. In doing so, we identified two more translational regulators, Brain tumor (Brat) and Argonaute-1 (Ago1), which physically interact with Bam and Sxl. Our results suggest a model in which translational repressors are assembled on the cis-element of nos mRNAs to mediate translational regulation.
MATERIALS AND METHODS
All chemicals were obtained from Sigma-Aldrich. Primary antibodies were obtained as follows; anti-FLAG (M2; Sigma) (mouse), anti-Myc (rabbit) (Cell Signaling), and anti-HA (rat) (Roche Applied Science). All secondary antibodies were obtained from Roche Applied Science.
The S2 cell expression vectors, pAc5.1A-FLAG- Bam and pAc5.1A-Myc-Bgcn were obtained from the previous studies (Kim et al., 2010). The NHL domain (aa 724- 1037), the N-terminus (aa 1-730), and the full-length of Brat (LD28374 from the Drosophila Genomics Resource Center) were cloned into the NotI/XbaI site of pAc5.1A-Myc to generate pAC5.1A-Myc-Brat NHL and a pAC5. 1A-Myc-Brat N terminal, and the pAC5.1A-Myc-Brat FL tagged proteins in S2 cells. For yeast two-hybrid, the full- length of Bam was cloned into Bam HI/XhoI sites of pLexA (Clontech) to generate LexA-Bam. The N- terminus and full length of Brat were cloned into the Bam HI/ SacI and Bam HI/XhoI, respectively, of pACT2 (Clontech) to generate TAD-Brat FL and -Brat N. The Brat NHL domain (aa 724-aa 1037) was cloned into SalI/Xba1 of pGAD 424 (Clontech) to generate TAD-Brat NHL. For the fragment complementation assay in HEK293T cells, the coding sequence and its derivatives of Bam and Brat were cloned into Bam HI/XhoI and the KpnI/HindIII site of the vectors (CoralHue_Fluo-chase kit; Medical and Biological Lab Co.), respectively, to produce protein fused to either the N- or the C- terminal fragment of the monomeric Kusabira-Green (mKG). P50- mKG_N and P65-mKG_C were provided as the control.
The various combinations of plasmids expressing the LexA DNA binding domain-fused protein and the GAD-fused protein were co-transformed into the yeast strain YPH500 (MAT α, ade 2, his3, leu 2, lys2, trp1, ura3) harboring the pSH18-34 plasmid (lexAop- LacZ reporter) (Gyuris, Golemis et al., 1993) by the standard lithium acetate method (Ito et al., 1983). The independent transformants were patched onto glucose plates containing X-gal for 24-48 hours, followed by a beta-galactosidase liquid assay as described (Ponticelli et al., 1995).
The Drosophila Schneider’s 2 cells (S2 cells) were maintained in a Shields and Sang M3 insect medium supplemented with 10% insect medium supplement (Sigma-Aldrich) and antibiotics (Hyclone) in a humidified atmosphere at 25℃. S2 cells were transiently transfected with expression vectors, using the DDAB method (Han, 1996). Transfected cells harvested 72h after transfection were washed in phosphate-buffered saline (Hyclone Dulbecco’s Phosphate Buffer saline) and lysed with radioimmune precipitation assay buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 5 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride; ELPIS-Biotech.) containing protease inhibitors. The lysates were clarified by centrifugation at 13,000 rpm (Eppendorf centrifuge) for 10 min at 4℃. The cleared extracts (3 mg) were mixed with 40 μL of anti-FLAG M2- conjugated agarose beads (Sigma Aldrich) and rotated at 4°C overnight. The beads were precipitated by Eppendorf centrifugation and washed three times with washing buffer (20 mM HEPES (pH 7.7), 150 mM NaCl, 2.5 mM MgCl2, 0.05% Nonidet P-40, 10% glycerol, and 1 mM dithiothreitol) containing protease inhibitors. The bound proteins were eluted in 50 μL of 0.1 mM glycine acetate (pH 3.0) and precipitated by adding 10% (the final concentration) trichloroacetic acid and 1% (final) sodium deoxycholate. The elutes were incubated 30 min at –20℃ and precipitated by centrifugation. The pellets were suspended in a 2XSDS loading buffer, and Western blot analysis was performed using anti-Flag and -Myc.
HEK293T cells were transiently transfected with vectors to express the mKG_N and mKG_C fusion proteins. The cells were plated on an 8-well cell culture slide (SPL) and then allowed to grow in medium. Then 24 h after transfection, 100 μL of 3.7% formaldehyde in 1XDPBS (Dulbecco's Phosphate Buffered Saline) was added to the cells for 5 min. The cells were then washed with 1XDPBS twice and were mounted on a drop of prolong Gold (Invitrogen). Images of the cells were acquired by confocal microscopy. GFP signals from either p50-mKG_N or p65-mKG_C alone or from a combination of these two were used as negative and positive controls, respectively.
RESULTS AND DISCUSSION
Genetic studies have shown that Bam is a critical factor in Nos repression and that its function requires Bgcn, Mei-P26, and Sxl (Chau et al., 2012; Li et al., 2013). However, individual interactions between them were not fully defined. We employed three different approaches to define these interactions.
To examine whether Bam physically interacts with Mei-P26, we performed a yeast two-hybrid assay (Kim et al., 2010). A LexA DNA binding domain (LexA) was fused to Bam full-length (FL) protein (LexA-Bam FL), while the transcriptional activation domain (TAD) of the Gal4 protein was fused to Mei- P26 FL protein. Transfection of yeast cells with LexA- Bam FL and TAD-Mei-P26 FL revealed that Bam physically interacted with Mei-P26. As Mei-P26 contains an NHL domain at its C-terminus, we tested whether that domain mediates interaction with Bam. Assays with the C-terminal NHL domain (Mei-P26 NHL) and the N-terminus (Mei-P26 N) lacking the NHL domain revealed that Bam interacted with the N- terminus (1-920), but not the NHL domain (921-1140) of Mei-P26 (Fig 1A).
Like Mei-P26, Brat is a member of the NHL domain protein family, is expressed in differentiating cells, and promotes stem cell differentiation (Harris et al., 2011), raising the possibility that Brat might function together with Bam in Nos repression. Thus, we performed a yeast two-hybrid assay to determine if Bam interacts with Brat, and found that LexA-Bam interacted with TAD-Brat FL. Unlike Mei-P26, LexA-Bam interacted with Brat NHL (724-1,037), but not the Brat N-terminus (1-730) (Fig. 1A).
We further performed pull-down assays to confirm the yeast two-hybrid results. Bam was fused to a Flag tag (Flag-Bam), while MeiP26 and its derivatives were fused to Myc tags (Myc-Mei-P26, Myc-Mei-P26 NHL and Myc-Mei-P26 N). Flag-Bam was then expressed with Myc-tagged proteins in S2 cells, and immunoprecipitation of S2 cell lysates performed with Flag antibodies. The Flag-Bam precipitates were subjected to western blot with Flag and Myc antibodies. Flag-Bam precipitates contained Myc- Mei-P26 and Myc-Mei-P26 N, but not Myc-Mei-P26 NHL (Fig. 1B), supporting results from the yeast two-hybrid assays. We conclude that that Bam physically interacts with the N-terminus of Mei-P26. Similar experiments were performed with Flag-Bam and Myc-Brat or Myc-Brat derivatives, confirming that Bam interacted with Brat through the Brat NHL domain (Table 1).
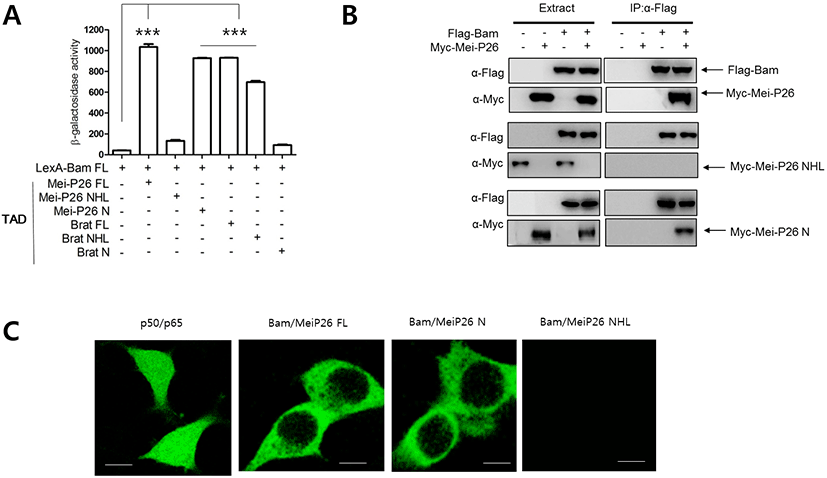
To further confirm our findings, we carried out GFP fragment complementation assays in mammalian cells (Kim et al., 2010). Bam was fused to an N-terminal fragment of GFP (Bam-GFP-N), while Mei-P26 and its derivatives were fused to the corresponding C-terminus fragment (Mei- P26-GFP-C, Mei-P26 N-GFP-C, and Mei-P26-NHL-GFP-C). Confocal microscopic observation revealed GFP fluorescence when Bam-GFP-N and Mei-P26 GFP-C fragments were co-expressed in cells. Neither single Bam-GFP-N nor single Mei-P26-GFP-C expression revealed GFP fluorescence (Fig. 1C). Bam-GFP-N and Mei-P26 N GFP-C, but not Mei-P26 NHL, did produce GFP fluorescence (Fig. 1C). Similar experiments were conducted with Bam-GFP-N and Brat-GFP-C, Brat N- GFP-C, or Brat-NHL-GFP-C fragments, confirming that Bam interacted with the Brat N-terminus, but not Brat NHL.
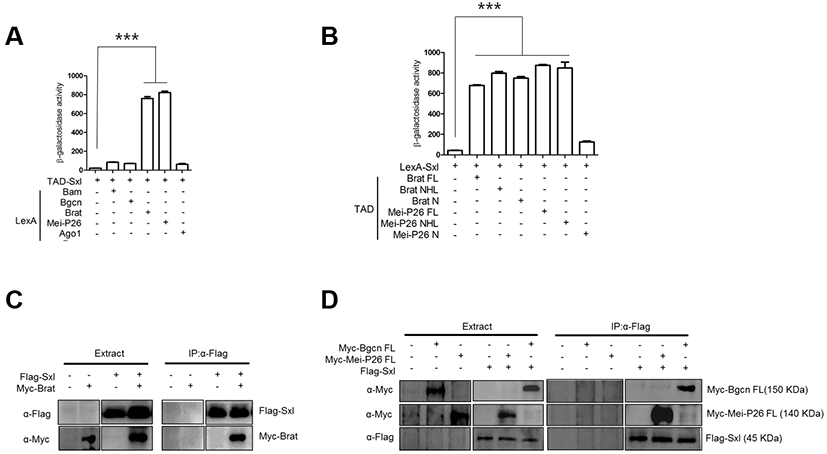
Like Bam, Sxl is a critical differentiation-promoting factor and represses Nos expression through interaction with the nos 3’UTR (Chau et al., 2012). Genetic analysis has shown that Bam requires Sxl for Nos repression (Li et al., 2013). We carried out yeast two-hybrid assays with TAD-Sxl and LexA-fused proteins, and found that Sxl did not interact with Bam or Bgcn (Fig. 2A). However, Sxl did interact with Brat and Mei-P26 (Fig. 2A). In particular, Sxl interacted with the C-terminal NHL domain but not the N- terminus of Mei-P26, and with both the Brat N-terminus and the Brat NHL domain. In pull-down assays, Flag-Sxl co-precipitated with Myc-Brat, Myc-MeiP26, and Myc-Bgcn.
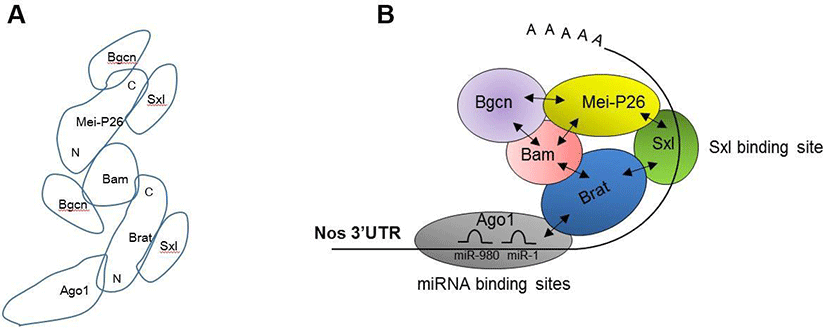
Our protein-protein interaction assays revealed a detailed map of interactions between Bam, Bgcn, Brat, Mei-P26, Sxl, and Ago1 (Fig. 3A). Based on this map, we propose a model illustrating how these proteins are assembled on the nos 3’UTR (Fig 3B). The model suggests that Sxl recognizes the distal part (781-850 3’UTR) of nos mRNAs, while Ago1-microRNA complexes recognize the proximal region (1-100 3’UTR). These two regions have been previously shown to mediate Nos repression (Li et al., 2009; Chau et al., 2012). Ago1 and Sxl recruit Brat, which in turn recruits Bam, Bgcn, and Mei-P26. These repressive complexes would mediate either translational repression or mRNA decay. Further research is needed to determine whether Ago1 and Sxl bind the proximal or distal regions of the nos 3’UTR, and whether binding of other translational regulators (Bam, Bgcn, Brat, Mei-P26) is dependent on Ago1 and Sxl. Moreover, it has yet to be determined whether Ago1 and Brat participate in Nos repression in vivo.