INTRODUCTION
Urechis species are marine animals belonging to the family Echiuridae, order Echiurida, and are widely distributed on the coast of the Yellow Sea, the southern sea of the Korean Peninsula, as well as in some parts of Japan, China, and the Pacific. In natural environments, Urechis spp. dwell in holes in a soft silt–clay sediment and gunk in the intertidal zone, where bivalves, crabs, and other crustaceans are abundant. Urechis spp. inhabit sandy mud caves of the U-type at certain depths. These echiurans are long- and soft-bodied animals. Despite their conservation and systematic diversity, little information is currently available regarding reproductive, biochemical, and environmental characteristics of Urechis species in Korea. Among Urechis spp., which can be discriminated reproductively (Choi et al., 1999; Shin & Kim, 2002), biochemically (Lee et al., 1998, 1999; Choi et al., 2017; Oh & Park, 2018), as well as environmentally (Choi et al., 1999, 2017; Kim, 2014), only few were examined at the genetic and/or molecular biological levels, unlike other invertebrates and/or teleost species. Thus, it is necessary to study genetic characteristics and the structure of echiuran populations to assess their genetic differences. Detailed information on genetic markers specific to lines, breeds, species, and/or geographical populations has been used for the identification and discrimination of individuals and populations, as well as hybrid pedigrees, and for molecular genetic testing (Mamuris et al., 1999; Klinbunga et al., 2000a; Nozaki et al., 2000; Yoon & Park, 2002; Kim et al., 2004; Park et al., 2005; Yoon & Kim, 2010; Yoon et al., 2012; Song & Yoon, 2013; Oh & Yoon, 2014). Accordingly, adaptations of PCR methods to the mariculture and fishing industry have been used to estimate genetic distances (GDs) among some finfish species and/or invertebrate animals from different geographical locations (Callejas & Ochando, 1998; Tassanakajon et al., 1998; Klinbunga et al., 2000b; McCormack et al., 2000; Yoon & Park, 2002; Kim et al., 2004; Park et al., 2005; Song & Yoon, 2013; Oh & Yoon, 2014). In this study, to evaluate the GDs and dissimilarities between geographical echiuran populations, clustering analysis of two Urechis spp. populations, from Gunsan and a Chinese coastal region, respectively, was performed.
MATERIALS AND METHODS
Experimental specimens were collected individually from Gunsan and Chinese Urechis spp. populations. Body muscle samples from 22 individuals, which were anesthetized with MS 222 (100 ppm), were taken from the body trunk and placed into pasteurized vials. The bodies of these Urechis spp. echiurans were collected into sterile tubes, which were immediately placed on ice and then stored in a refrigerator until use. DNA extraction was performed as described previously (Oh & Yoon, 2014). Chloroform (500 μL) was added to the mixture, and the content of the tube was mixed by inversion. After washing, samples of body muscle tissues were placed into 10-mL test tubes, to which three volumes of lysis buffer were added, and the content of the tubes was mixed by gentle inversion. The precipitates collected were incubated with 20 μL of a proteinase K solution (15 mg/mL). After the incubation, 300 μL of 6 M NaCl was added, and the mixture was gently pipetted for a few minutes. The DNA pellets were dried for 3 h, then stored at −90℃ until use, and thawed in TE buffer. The purified genomic DNA was quantified by measuring absorbance at 260 nm using a spectrophotometer (Beckman Coulter, Buckinghamshire, UK). The DNA tablets were then dried for more than 10 h, stored at −90℃ until use, and thawed in purified DW.
Oligonucleotides primers were purchased from Operon Technologies, USA. OPA-17 (5′-GACCGCTTGT-3′), OPB-20 (5′-GGACCCTTAC-3′), OPC-01 (5′-TTCGAGCCAG-3′), OPC-04 (5′-CCGCATCTAC-3′), OPC-09 (5′-CTCACCGTCC-3′), and OPD-05 (5′-TGAGCGGACA-3′) were used to reveal unique loci to each population and shared loci between the two Urechis spp. populations. These oligonucleotides primers were used to determine genetic variations, DNA polymorphisms, and pairwise similarities between Urechis spp. from Gunsan and the Chinese location. PCR was performed using a programmable DNA thermal cycler (MJ Research, Inc., Waltham, MA, USA). DNA amplification was carried out in 25-μL reactions, which contained 10 ng of template DNA, 22 μL of a premix (Bioneer Corp., Daejeon, Korea), and 1 unit of a primer. Amplified fragments were separated by electrophoresis in agarose (Bioneer Corp.) gels with TBE using a 100-bp DNA ladder (Bioneer Corp.) as a DNA molecular weight marker, followed by staining with ethidium bromide. The electrophoresed agarose gels were illuminated with UV light, and photographs were taken using a PhotoMan direct copy system (PECA Products, Beloit, WI, USA).
A similarity matrix comprising band-sharing (BS) values was designed, along with the presence/absence of amplified fragments at specific spots in identical gels from DNA frameworks. BS values were estimated according to Jeffreys & Morton (1987) and Yoke-Kqueen & Radu (2006). Based on comparison of two lanes, the BS value was calculated as follows: BS=2 (Nab)/(Na+Nb), where Nab is the number of fragments shared by samples a and b; Na is the total number of fragments in sample a; and Nb is the total number of fragments in sample b. The average within-population similarity was assessed by pairwise comparison between individuals within a population. The relationship between unlike individuals in the Urechis spp. populations was estimated, along with the BS values and similarity matrix. A hierarchical polar dendrogram was constructed using similarity matrices to produce a group tree using Systat version 10 (SPSS, Inc., Chicago, IL, USA). The Systat software was also used to evaluate GDs, Euclidean GDs within and between the Urechis spp. populations, means, standard errors, and t-test values.
RESULTS AND DISCUSSION
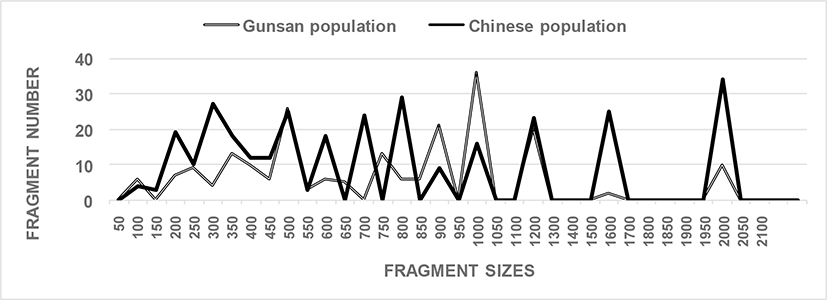
In this study, six oligonucleotides primers, OPA-17, OPB-20, OPC-01, OPC-04, OPC-09, and OPD-05, were utilized to identify unique loci to each population and shared loci between the two Urechis spp. populations. The complexity of the banding patterns varied significantly between primers to two regions. The number of fragments in each size interval was computed by combining fragments obtained with all six oligonucleotides primers, as shown in Fig. 1. The sizes of the DNA fragments also varied greatly, from 50 to 2,050 bp, as displayed in Fig. 1. Fragments >2,100 bp were not detected in the two Urechis spp. populations. A cluster tree for the two Urechis spp. populations was built using UPGMA Euclidean GD analysis based on 543 fragments generated by using the six oligonucleotides primers. In finfish, a phylogenetic tree was constructed using UPGMA cluster analysis based on 3,744 distinguishable fragments from gynogenetic clones of the silver crucian carp, Carassius auratus gibelio Block (Zhou et al., 2000). The sizes of fragments varied from 220 to 1,700 bp in four species from the Mullidae family (Mamuris et al., 1999). DNA fragments obtained using four primers ranged from 100 to 2,300 bp in the brittle star (Amphiura filiformis) (McCormack et al., 2000). In this study, the average number of distinct fragments was 25.67 per primer in the Chinese echiuran population, while the number of recorded fragments varied from 7 to 12 per primer in four species from the Mullidae family (Mamuris et al., 1999).
The BS value between individuals no. 05 from the Gunsan population and no. 22 from the Chinese population of Urechis spp. was 0.206, which was the lowest observed value, as illustrated in Table 1. The BS value between individual’s no. 17 and no. 18 was 0.939, which was the highest value, discovered within the Chinese Urechis spp. population. Twenty-two unique loci were found in the Gunsan Urechis spp. population using the OPA-17 oligonucleotides primer, as summarized in Table 2. Remarkably, oligonucleotides primers OPC-04 and OPB-20 revealed 44 and 33 unique loci in the Chinese Urechis spp. population, respectively (Table 2), which were able to distinguish the two populations. Notably, primer OPB-17 allowed the detection of 22 shared loci between the two populations, which were present in all samples, as summarized in Table 2. Oligonucleotides primer OPD-05 also allowed the discovery of 22 shared loci between the two Urechis spp. populations, with major and/or minor fragments of 1,200 bp present in all samples. The average number of unique loci was found to be 2.5 times higher in the Chinese than in the Gunsan population. A total of 271 unique loci, with an average of 38.7 loci per primer, were identified in the GSCP population. Furthermore, 33 loci, with an average of 4.7 loci per primer, were found to be shared by three hard clam (Meretrix lusoria) populations (Yoon et al., 2012). A decamer primer, BION-B14, revealed seven unique loci, with fragments of approximately 100 bp, which were able to distinguish each population of Vietnamese cuttle fish (Sepia esculenta) (Yoon & Kim, 2010). Remarkably, 21 unique loci were detected using the BION-B06 primer, with fragments of approximately 100, 450, and 650 bp, in a Sockcho cuttlefish population.
The average BS value was lower (p<0.05) for individuals from the Gunsan population (0.661±0.012) than for those from the Chinese population (0.788±0.014), as demonstrated in Table 3. Meanwhile, the average BS value in this study was higher than that between the common carp and Israeli carp species (0.57±0.03) (Yoon, 2001) and in a Geojedo oyster population (0.537±0.017) (Kim et al., 2004). However, the average BS value in this study was similar to that in the Spanish barbel species (0.71–0.81) (Callejas & Ochando, 1998). In terms of average BS values, a cuttlefish (S. esculenta) population from Sockcho exhibited a higher BS value (0.826) than did fish from Seocheon (0.465) (Yoon & Kim, 2010). Polymorphisms are determined based on banding patterns of primer-amplified products at specific sites (Mamuris et al., 1999; Klinbunga et al., 2000a; Nozaki et al., 2000; Yoon, 2001; Yoon & Park, 2002; Kim et al., 2004; Oh & Yoon, 2014). Several studies have reported the sizes of PCR fragments for black tiger shrimp (Penaeus monodon) (Tassanakajon et al., 1998), the brittle star (A. filiformis) (McCormack et al., 2000), and lobster species (Park et al., 2005). Unambiguous data, resulting from variations in DNA polymorphisms, were found to be meaningful in the identification of individuals and/or populations (Tassanakajon et al., 1998; Mamuris et al., 1999; Yoon & Park, 2002; Chenyambuga et al., 2004; Kim et al., 2004; Yoon & Kim, 2010; Yoon et al., 2012; Song & Yoon, 2013).
| Population | Gunsan | Chinese |
|---|---|---|
| Gunsan | 0.661±0.012 a | 0.304±0.007 b |
| Chinese | - | 0.788±0.014 c |
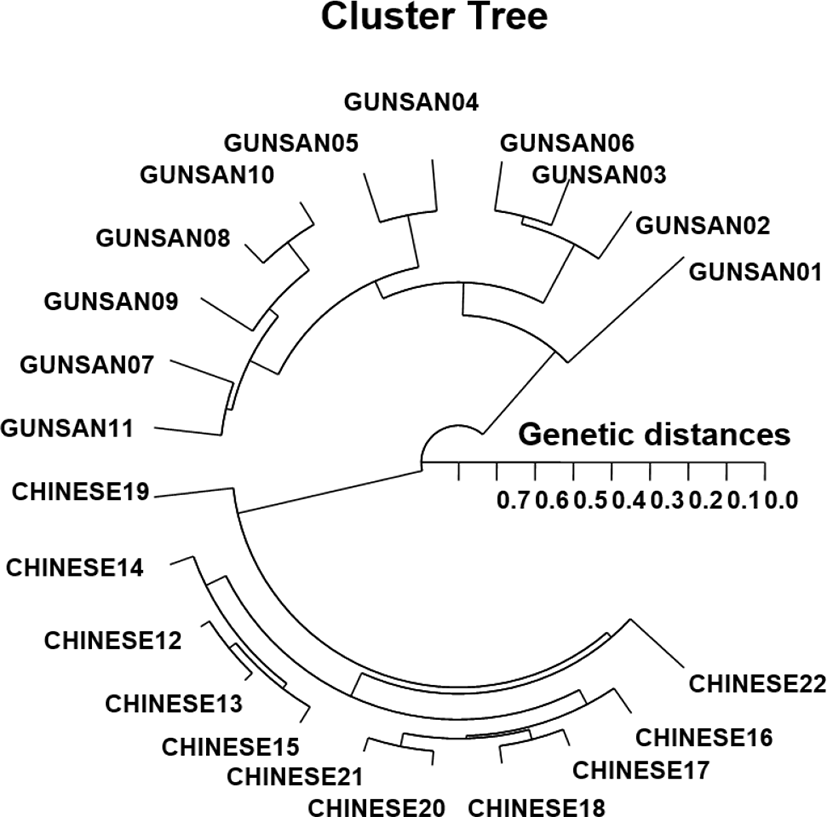
In this study, PCR led to the discovery of a significant GD between the two populations/species. PCR enabled the discrimination of the populations/species and the detection of genetic variations within the two geographical populations of Urechis spp. The hierarchical polar dendrogram delineated two main groups, group 1 (GUNSAN 01–11) and group 2 (CHINESE 12–22), within the two geographical Urechis spp. populations. The shortest GD (0.027), exhibiting a significant molecular difference, was found between individuals CHINESE no. 12 and no. 13. Meanwhile, individual GUNSAN no. 06 of Urechis spp. was most distantly related to CHINESE no. 22 (GD=0.703). These results demonstrated that the Gunsan Urechis spp. population was genetically different from the Chinese Urechis spp. population. A neighbor-joining tree constructed based on GDs among populations, using a PCR method, was used to clarify the relationships between three mud crab species (Klinbunga et al., 2000b). The results showed that there were great genetic differences between geographically separated populations within a species and between species. Phylogenetic relationships among five Haliotis species and one hybrid were elucidated by calculating distance coefficients and constructing a phylogenetic tree based on PCR data (Kim et al., 2000). These species were divided into two clusters, of which cluster I included H. discus hannai, H. discus, H. gigantea, H. sieboldii, and the hybrid and was further divided into two subclusters. Subsequently, these authors reported that PCR was a preferred tool for determining phylogenetic relationships among six Haliotis species.
As mentioned above, the prospect of PCR analysis to detect diagnostic markers for the identification of the two Urechis spp. populations was recognized, confirming that the technique is a suitable approach for DNA assessments, both within and between individuals, populations, and species. The classification of echiuran species is based on morphological variations in the body size, body type, body color, body wall, proboscis type, and proboscis size. It has been presumed that differences in these traits between species reflect their separate origin or genetic identity (Chenyambuga et al., 2004). Information on GDs among echiurans would be beneficial to comprehend the expansion or maintenance of individual populations in inshore and tidal flat regions of the Korean Peninsula. However, additional research, using more individuals, populations, species, and genera, as well as more primers, will be needed to fully elucidate the specificity of loci to definite taxa and subsequently explore the interspecific gene flow in the genus Urechis. Further studies including larger numbers of samples and primers need to be conducted to obtain more detailed information on the genetic structure of the two species of echiurans.

